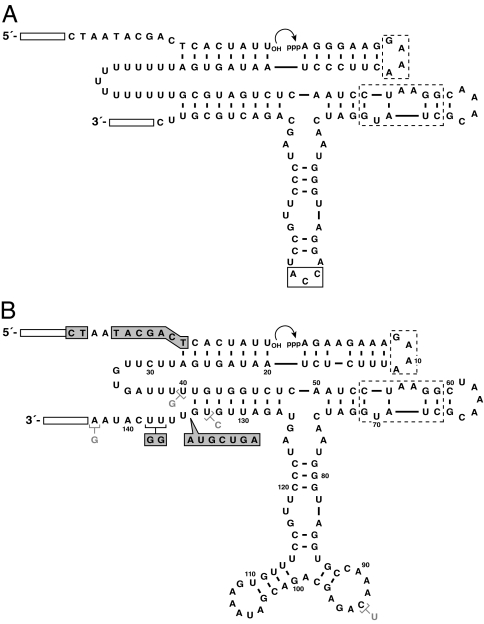
Fig. 2.
Sequence and secondary structure of the starting and evolved DSL ribozymes. Open rectangles indicate primer binding sites, which are immutable during the course of evolution. Dashed boxes indicate the tetraloop and tetraloop receptor. (A) Modified DSL ligase that was used as the basis for constructing the starting pool of ribozymes. Boxed residues were replaced by 35 random-sequence nucleotides. (B) T80-1 ligase, which was obtained after transfer 80 of continuous evolution. Ribozyme residues in shaded boxes indicate changes present in all clones obtained after transfer 100. These changes confer complementarity to substrate residues shown in shaded boxes. Additional mutations present in the T100-1 ligase are shown in gray.