Fig. 2.
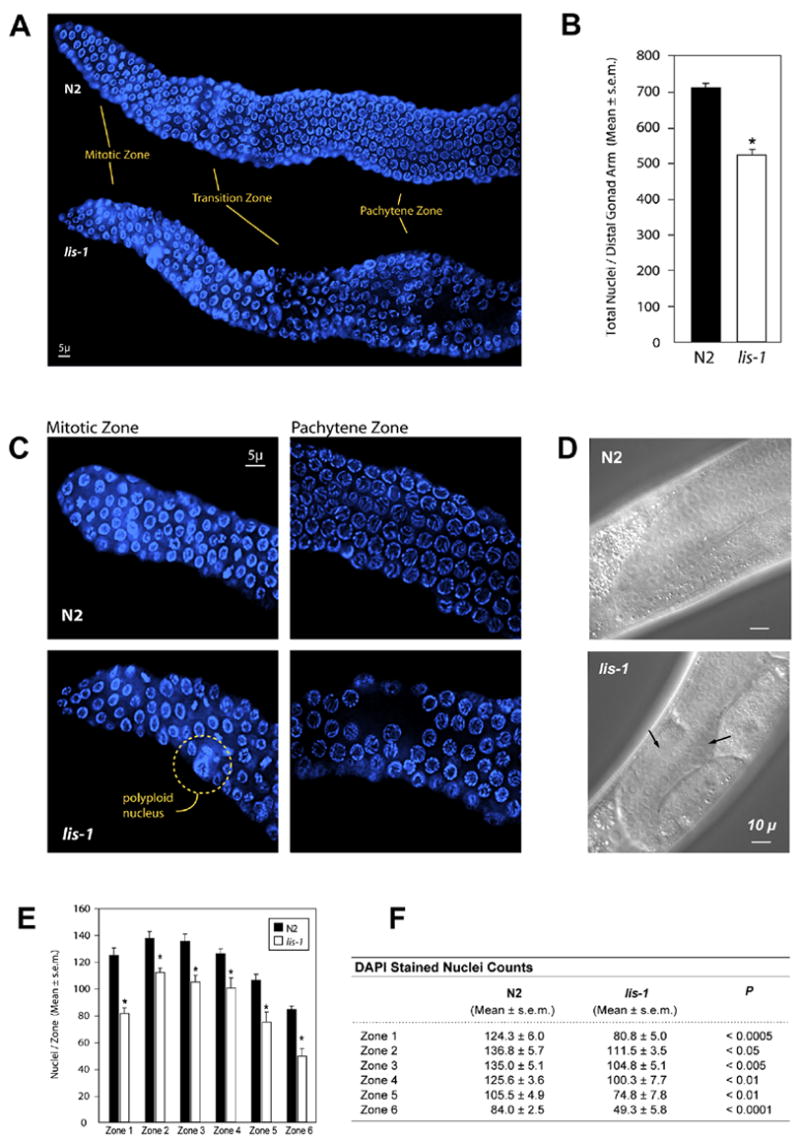
lis-1(lf) disrupts development throughout the germline. (A) Hoechst staining reveals defects in the mitotic, transition, and pachytene zones of lis-1(lf) distal gonads. (B) lis-1(n3334) mutants contain fewer germ cells per distal gonad arm than the N2 strain. lis-1 homozygotes were identified by selecting GFP-negative progeny of strain MT12272. The data are averages ± s.e.m. for N2 (n=8) and lis-1 (n=4) animals. N2 distal gonad arms contain an average of 711.1 ± 12.4 nuclei, whereas lis-1 distal gonad arms contain an average of 521.3 ± 18.1 nuclei (P<0.0001, unpaired t-test). (C) The mitotic zone of lis-1(n3334) mutants contains polyploid nuclei. The pachytene zone of lis-1(n3334) mutants contains large cell-sparse gaps. (D) Arrows point to cell-sparse gaps in the lis-1 germline visible by Nomarski DIC. (E, F) We counted germ cells in 40 μm zones from the distal tip (Zone 1) to the bend of the gonad (Zone 6). Cell counts were significantly lower in each zone of the lis-1(n3334) distal gonad than in the corresponding N2 zones.
