Figure 2.
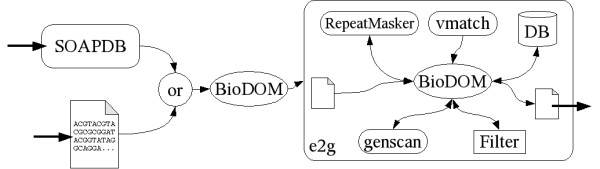
Graph representation of the e2g workflow. The e2g webservice gets sequence information in SequenceML format (besides a couple of parameters) as input and returns the result data as an EBIApplicationResult XML document. The input data can originate from a file (containing sequence information as SequenceML) or from an external data source like the SOAPDB webservice (which returns sequence information in FASTA format). Using BioDOM as a converter between the different data formats, it is quite easy to add another data source. The e2g webservice is a workflow itself and also uses webservice technology to mask repeats (using the RepeatMasker webservice) and match the input sequence data against huge EST databases (using the vmatch webservice). The match result is filtered (depending on input parameters) and returned as an EBIApplicationResult document.
