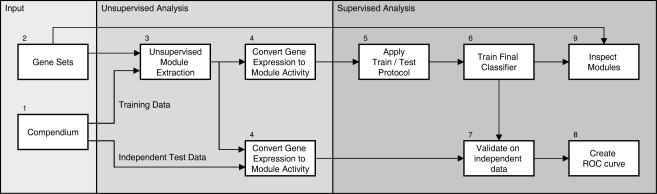
Figure 1. Workflow of the approach.
We extended the analysis of compendia [10] to the supervised classification domain. Several microarray datasets were collected to construct compendia at various levels of underlying phenotype diversity (1). Additionally, we gathered a collection of biologically meaningful gene sets from available databases (2). Using the module extraction framework proposed by [10], we derived sets of modules (3) from these compendia and gene sets. Using these modules we construct a module activity matrix (4), allowing modules rather than single genes to be used as features. The predictive power of the different sets of modules is inspected within a classification context. Using a train/test protocol (5), we estimated the generalization error of all sets of modules [17]. Succeedingly, we trained a final classifier (6), which was then validated on independent data (7), and its performance assessed (8). Furthermore, the approach allows the final set of modules that were selected in the classifier to be compared to the original gene sets (9), allowing the identification of biological processes underlying the development and progression of cancer.