Abstract
Jumonij (JMJ)/Jarid2 plays important roles in embryonic development and functions as a transcriptional repressor. Using yeast two-hybrid screening, we have identified a cofactor of JMJ, the zinc finger protein 496 (Zfp496) that contains a SCAN, KRAB and zinc finger domain. Our molecular analyses indicate that Zfp496 functions as a transcriptional activator. Further, Zfp496 inhibits the transcriptional repression of JMJ and JMJ represses the transcriptional activation of Zfp496. This study demonstrates that JMJ physically and functionally interacts with Zfp496, which will provide important insights into endogenous target gene regulation by both factors.
Keywords: jumonji, zfp496, transcription, heart, development
INTRODUCTION
Significant advances have recently been made in understanding the role of transcription factors during embryonic development including heart development. Transcription factors that play critical developmental roles in the heart belong to many different transcription factor families. These include, but are not limited to, the homeodomain protein Nkx2.5 [1], the zinc finger protein GATA4 [2,3], the MADS box factor MEF2C [4,5], and the T-box transcription factor Tbx5 [6]. Loss of these transcription factors in mice results in early embryonic lethality due to cardiac defects. Increasing evidence indicates that complex interactions among multiple trans-acting factors positively or negatively regulate the target gene expression. Nkx2.5 activates expression of atrial natriuretic factor (ANF) by binding to the enhancer region [7,8], and interacts cooperatively with other transcription factors including GATA4 and PITX2C [9]. GATA4 interacts with FOG-2 (friend of GATA), which results in repressing GATA4 activities [10]. Tbx5 synergistically activates the ANF promoter with GATA4 and Nkx2.5 [11], while Tbx20 represses the ANF activation mediated by Tbx5 [12].
By searching for developmentally important genes, the jumonji gene (jmj) was identified as a critical nuclear factor for mouse embryonic development including cardiovascular development [13,14] (for review see [15]). Jmj has recently been renamed Jarid2 according to NCBI nomenclature but will be referred to in this manuscript as jmj. JMJ belongs to the AT-rich interaction domain (ARID) transcription factor family [16-18], and is a member of the Jumonji transcription factor family [19,20]. Recently, members of the Jumonji transcription factor family including JHDM1 [21], JHDM2A [22], and JMJD2A [23] have been shown to exhibit histone demethylase activity. However, the function of JMJ as a histone modifier remains to be elucidated. We have previously reported that knockout of jmj results in heart malformations, which mimic human congenital heart diseases (for review see [15]). Our structural/functional analyses of JMJ indicate that JMJ functions as a transcriptional repressor and inhibits ANF activation by Nkx2.5 and GATA4 [24,25]. JMJ is involved in regulating cardiomyocyte proliferation in a temporal- and spatial-dependent manner [26,27]. At the molecular level, JMJ represses cardiomyocyte proliferation through interaction with the retinoblastoma protein [28].
Although the functional roles of JMJ are emerging in association with embryonic development, the molecular mechanisms by which JMJ functions remain largely unknown. Therefore, we performed a yeast two-hybrid screen to identify novel cofactors of JMJ, which would lead us to determine critical factors for target gene regulation. Using JMJ as bait, a zinc finger protein 496 (Zfp496) was identified whose function remains unknown. Zfp496 consists of three distinct domains. The SCAN domain, which is also called LeR because it is a leucine-rich region, is a protein interaction domain that mediates both self-association and association with other SCAN-box proteins. A KRAB domain functions as a transcriptional repressor when tethered to the template DNA by a DNA-binding domain (for review see [29]). Zinc finger regions of proteins are thought to interact directly in both protein-DNA interactions and protein-protein interactions. Zfp496 contains 5 zinc finger regions, of which four are conventional C2H2 fingers, while one is a novel C2HR motif [30]. KRAB-zinc finger proteins probably constitute the single largest class of transcription factors within the human genome [31]. Nielsen et al. [30] recently reported Zfp496 as Nizp1 (NSD1-interacting zinc finger protein) while we were characterizing the function of this protein. NSD1 (Nuclear receptor-binding SET-domain containing protein), the histone lysine methyltransferase, was initially identified to interact with retinoic acid receptor and exhibited characteristics of both corepressors and coactivators [32].
In this study we show that JMJ physically interacts with Zfp496 in vivo and in vitro. Interestingly, Zfp496 functions as a transcriptional activator when it is tethered to DNA or by directly binding to the DNA binding motif. Further we show that JMJ and Zfp496 are able to attenuate each other’s transcriptional activity. The complex interactions among transcription factors are the key regulatory components of the temporal- and spatial-expression of target genes. These findings will provide the important basis to further study the endogenous gene regulation by JMJ and Zfp496.
MATERIALS AND METHODS
Yeast two-hybrid screening
To identify cofactors that interact with JMJ, a yeast two-hybrid screen was performed using a bait containing the full length JMJ. A mouse embryonic expression library was screened according to the manufacturer’s recommendation (OriGene Technologies, Inc). Putative positive library plasmids were recovered from yeast and subjected to confirmation mating experiments by streaking the two yeast strains containing a bait or a potential positive cDNA. For positives that passed confirmation tests, the cDNAs were identified by sequencing and Blasting against the NCBI GenBank sequence. A total of 25 independent positive clones were recovered and sequenced. Among several potential cofactors selected according to our criteria such as nuclear localization and putative transcription activity, Zfp496 was further characterized as a potential JMJ cofactor in this study.
Plasmid constructs
The recovered cDNA from yeast contained nucleic acids 1590-2375 of Zfp496 that encodes amino acids (aa) 450-585 when compared to the published sequence (GeneBank accession number gi25955548). The full length coding sequence of Zfp496 was obtained by RT-PCR amplification with gene specific primers, and verified by sequencing. Zfp496 mutant constructs in the pcDNA3.1/myc-His (-) B (Invitrogen), GAL4-DNA-binding domain (DBD)/pcDNA3.1/Xpress vector, and the glutathione S-transferase (GST)-Zfp496 plasmid (pGEX-2T, Amersham) were generated as previously described [24]. All JMJ constructs used were previously described [24]. The reporter gene pG5Ti consisted of 5X GAL4 binding sites upstream of the adenovirus major late promoter TATA box linked to luciferase (obtained from Dr. P. Farnham) [33]. SAAB reporter constructs were generated by tandem ligation of the SAAB oligonucleotides followed by subcloning into pcRII-TOPO vector (Invitrogen). The oliognucleotide inserts were excised using XhoI and SacI and ligated into a pGL2 vector (Promega) containing an E1B promoter (obtained from Dr. Eric Olson). The ligated sequences for the reporter constructs are: pGL2-E1B-4X SAAB #3 5’-CTATTACTTGTAT-3’; pGL2-E1B-5X SAAB #4 5’-ATTAATTATTGATCA-3’; and pGL2-E1B-5X SAAB #18 5’-AACTGGCCATCCAGC-3’.
Protein-protein interaction
To examine the in vivo association of Zfp496 with JMJ, coimmunoprecipitation was performed as previously described [8,28,30] with minor modifications. These modifications include preclearing the nuclear extract by incubation with mouse immunoglobulin G (IgG) and protein G-agarose (Sigma) for 1 hour before 3 μg of anti-myc (Santa Cruz Biotechnology) or anti-FLAG (Sigma) antibody was added. The immunoprecipitates were analyzed by Western blotting with anti-myc or anti-FLAG antibody. For a semi in vivo approach to the coimmunoprecipitation, heart extracts from 7 day old mice were prepared using a buffer containing 0.2% SDS, 11% glycerol, 11mM Tris [pH 8.0], 1.1 mM EDTA [pH 8.0], 1mM DTT and protease inhibitors. Hearts were homogenized and then subjected to sonication before a final centrifugation at 15,000 × g at for 30 minutes. 2 mg of total protein extract was diluted 1:10 in NETN buffer (20mM Tris-HCI (pH8.0), 100mM NaCl, 1mM EDTA, and 0.5% NP-40) containing 1mM DTT and protease inhibitors. The extract was pre-cleared with GST beads and then incubated at 4° overnight with 5 μg of GST-Zfp496 beads or GST-beads. The bound proteins were subjected to Western blotting using a JMJ-P100 antibody [24].
To map the protein-protein interaction domains between Zfp496 and JMJ, GST pull-down assays were performed [8] using the TNT T7 Coupled Reticulocyte Lysate System (Promega). 35S-methionine labeled proteins were incubated in NETN buffer with GST-glutathione beads, GST-Zfp496 beads, GST-J-Nt (N-terminal, aa 1-528) beads or GST-J-Ct (C-terminal, aa 529-1234) beads. 35S-labeled FL Zfp496 consists of aa 1-585, 35S-Zfp496 SCAN only consists of aa 1-198, 35S-Zfp496 KRAB only consists of aa 155-333, and 35S-Zfp496 zinc only consists of aa 333-585. For the reciprocal experiment, 35S-FL JMJ (aa 1-1234), 35S-JMJ aa 1-220, 35S-JMJ aa 1-130, and 35S-JMJ aa 131-222 were used.
Reporter gene assay, Gel mobility shift assay (GMSA), and Immunostaining
10T1/2 cells were transfected using calcium phosphate precipitation method [8], or Lipofectamine 2000 (Invitrogen). Data are given as means ± standard error of the mean. Statistical analyses were performed using the student’s paired t-test. Results were accepted as significant when p was <0.05, which is indicated by *. To examine whether Zfp496 binds to DNA in vitro, GMSAs were performed as described previously [34]. Indirect immunostaining experiments were described elsewhere [13].
Site selection and amplification of binding (SAAB)
To identify the DNA-binding consensus sequences of Zfp496, the SAAB method was carried out as described previously [24,35,36] with minor modifications. A random sequence library was prepared by synthesizing oligonucleotide pools comprising a random 15-bp sequence flanked by 18 base linker sequences 5′-GCGAAGCTTTGACTGAGG(N15)TTGATGCCGAGGATCCCG-3′ (Integrated DNA Technologies). BamHI and HindIII sites are underlined. The PCR primers annealing to the 18 bases of each top and bottom strand were synthesized (5’ sense: 5’-GCGAAGCTTTGACTGAGG; 3’ anti-sense: 5’-CGGGATCCTCGGCATCAA). A primer extension reaction was performed to make double-stranded, 32P-labeled oligonucleotides, which were purified by PAGE. 10 μg of GST-Zfp496 was incubated with 1 μg of poly (dI-dC) in the Reaction Buffer (10 mM Tris, pH 8.0, 1 mM dithiothreitol, 1 mM NaH2PO4, 5% glycerol, 50 mM NaCl, and protease inhibitors) for 10 minutes on ice. 2 μg of 32P-labeled random double-stranded probe was added for 20 minutes at room temperature and subjected to GMSA. After the shifted band was excised, the DNA was eluted from the gel and amplified using 15 PCR cycles. The selection procedure was repeated for four rounds. After the last round of selection, PCR-amplified products were directly subcloned into the pCRII-TOPO vector and sequenced. These oligonucleotides were then subjected to GMSA to confirm their binding to Zfp496.
RESULTS
Zfp496 physically associates with JMJ in vivo
In this study, Zfp496 was identified as a potential cofactor of JMJ using a yeast two-hybrid screen. The recovered cDNA from yeast contained the last two zinc finger motifs, suggesting that this zinc finger region of Zfp496 is sufficient to interact with JMJ in yeast. To investigate whether Zfp496 associates with JMJ in vivo, coimmunoprecipitation experiments were performed using HEK 293 cells overexpressing Zfp496 and JMJ (Fig. 1). As shown in Figure 1A, Zfp496-myc protein was detected (lane 2) when the cell extract coexpressing FLAG-tagged JMJ (JMJ-FLAG) was immunoprecipitated with anti-FLAG antibody, followed by Western blotting with anti-myc antibody. Zfp496 was not detected when the control cell extract expressing only Zfp496-myc was used (lane 1), indicating the specific interaction of JMJ with Zfp496. The converse coimmunoprecipitation experiment also revealed the specific interactions of Zfp496 with JMJ in vivo (Fig. 1B). When the cell extract containing Zfp496-myc and JMJ-FLAG was immunoprecipitated with anti-myc antibody, Zfp496 was able to pull down JMJ (lane 2). JMJ was not detected when coimmunoprecipitation was performed with anti-myc antibody in the cell extract containing only JMJ-FLAG (lane 1). These data clearly demonstrate that Zfp496 physically associates with JMJ in vivo when the proteins are coexpressed.
Figure 1.
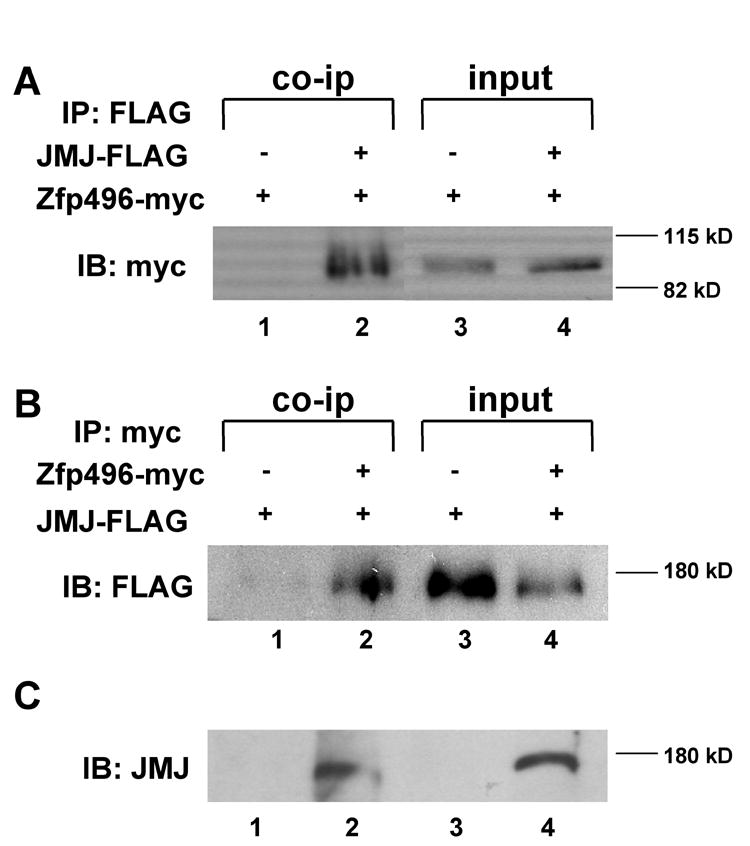
Zfp496 physically interacts with JMJ in vivo. Coimmunoprecipitation was performed using HEK293 cells transected with FLAG-tagged JMJ and myc-tagged Zfp496. A. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody followed by immunoblotting (IB) with anti-myc antibody. Lanes 3 and 4 represent direct western blotting of the cell lysates and are 4% of the input of co-ip. B. The reverse experiment was done to confirm the protein interaction. Cell lysates were immunoprecipitated with anti-myc antibody followed by immunoblotting with an anti-FLAG antibody. Lanes 3 and 4 represent direct western blotting of the cell lysates and are 4% of the input of the co-ip. C. Semi in vivo GST-pull-down assay. Protein extract from mouse heart was incubated with GST beads (lane 1) or GST-Zfp496 beads (lane 2) followed by Western blotting using the JMJ antibody. Direct Western blotting of 10T 1/2 control cells (lane 3) or heart extract (lane 4) is shown as controls.
To further show the in vivo interaction between Zfp496 and JMJ, a semi in vivo GST-pull-down experiment was performed using heart extracts. As shown in Fig. 1C, this result clearly shows that GST-Zfp496 is able to interact with endogenous JMJ expressed in the heart. The JMJ band was detected when heart extracts were incubated with GST-Zfp496 (lane 2), but not with GST alone (lane 1). Direct Western blotting shows the single JMJ band in the heart extract (lane 4), but not in the 10T1/2 control cells (lane 3). The reciprocal experiment could not be performed due to lack of a suitable Zfp496 antibody.
The N-terminal of JMJ interacts with the zinc finger region of Zfp496 in vitro
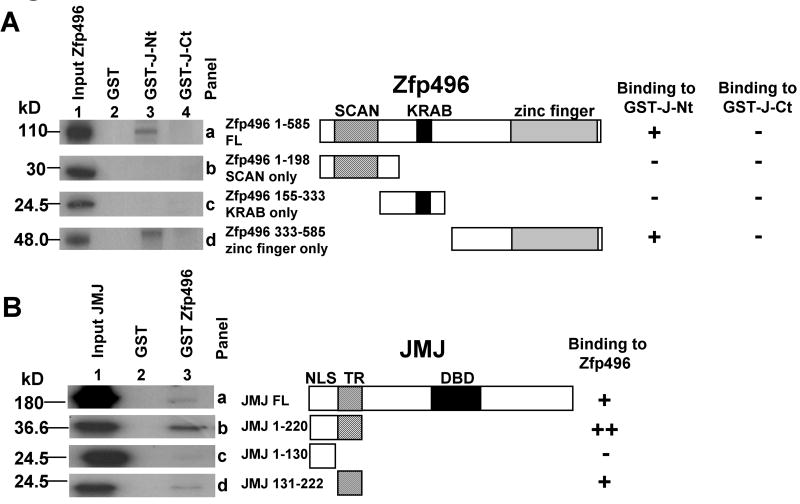
To investigate the regions involved in the protein-protein interaction between Zfp496 and JMJ, GST pull-down assays were performed (Fig. 2). The JMJ protein contains several domains which have been previously described [24]. The nuclear localization signal (NLS) of JMJ is present between aa 1-131. The region responsible for transcriptional repression (TR) is comprised of aa 132-222 [24]. JMJ-C-terminal mutant (J-Ct, aa 529-1234) consists of the JmjN, ARID, and JmjC domains, which includes the DNA-binding domain (DBD). The JMJ-N-terminal mutant (J-Nt) consists of aa 1-528. 35S-labeled full length (Fig. 2A, panel a) and Zfp496 mutants (panels b, c and d) (lane1) were incubated with GST (lane 2), GST-J-Nt (lane 3), or GST-J-Ct (lane 4) coupled to agarose beads. Full length-Zfp496 was able to bind to the GST-J-Nt but not GST-J-Ct (panel a). Neither Zfp496-SCAN only mutant (aa 1-198, panel b) nor the Zfp496-KRAB only mutant (aa 155-333, panel c) was able to bind to GST-J-Nt or GST-J-Ct. Zfp496-zinc only (aa 333-585, panel d) did bind to GST-J-Nt but not GST-J-Ct. This assay indicates that Zfp496 and JMJ can interact in vitro and the interaction is between the N-terminal region of JMJ and the zinc finger region of Zfp496, which is consistent with the result of yeast two-hybrid screening.
Figure 2.
Mapping of protein interaction sites in vitro. A. In vitro 35S-Zfp496-full length (FL) and mutants were incubated with GST, GST-J-Nt (aa 1-528) or GST-J-Ct (aa 529-1234) linked to agarose beads. The bound proteins were resolved using SDS-PAGE followed by autoradiography. Panels a through d are: Zfp496-FL (aa 1-585), Zfp496-SCAN only (aa 1-198), Zfp496-KRAB only (aa 155-333), and Zfp496-zinc finger only (aa 333-585), respectively. B. In vitro 35S-FL-JMJ and JMJ mutants were incubated with GST or GST-Zfp496 linked to agarose beads. Panels a through d are: FL-JMJ aa 1-1234, JMJ aa 1-220, JMJ aa 1-130 (NLS), and JMJ aa 131-222 (TR), respectively. The DNA-binding domain of JMJ (DBD, aa 529-792) is indicated. (-, no interaction; +, interaction; ++, strong interaction).
To perform the reciprocal experiment (Fig. 2B), 35S-JMJ constructs (lane 1) including JMJ full length (FL, panel a), JMJ 1-220 (panel b), JMJ 1-130 (panel c), and JMJ 131-222 (panel d) were incubated with GST (lane 2), or GST-Zfp496 (lane 3) coupled to agarose beads. GST-Zfp496 associated with full length JMJ as well as with JMJ 1-220 and to a lesser extent JMJ 131-222. Zfp496 did not associate with JMJ 1-130, indicating that Zfp496 interacts with the TR domain of JMJ and that the TR domain of JMJ is sufficient for the interaction. The results indicate that the zinc finger region of Zfp496 interacts with the TR domain of JMJ in vitro. For all GST-pull-down experiments, GST alone did not bind to any of the 35S-proteins indicating specificity of the protein interactions between JMJ and Zfp496. Other 35S-JMJ-C terminal proteins such as JMJ aa 529-792 were tested in the assay and showed no interaction (data not shown).
Zfp496 functions as a transcriptional activator
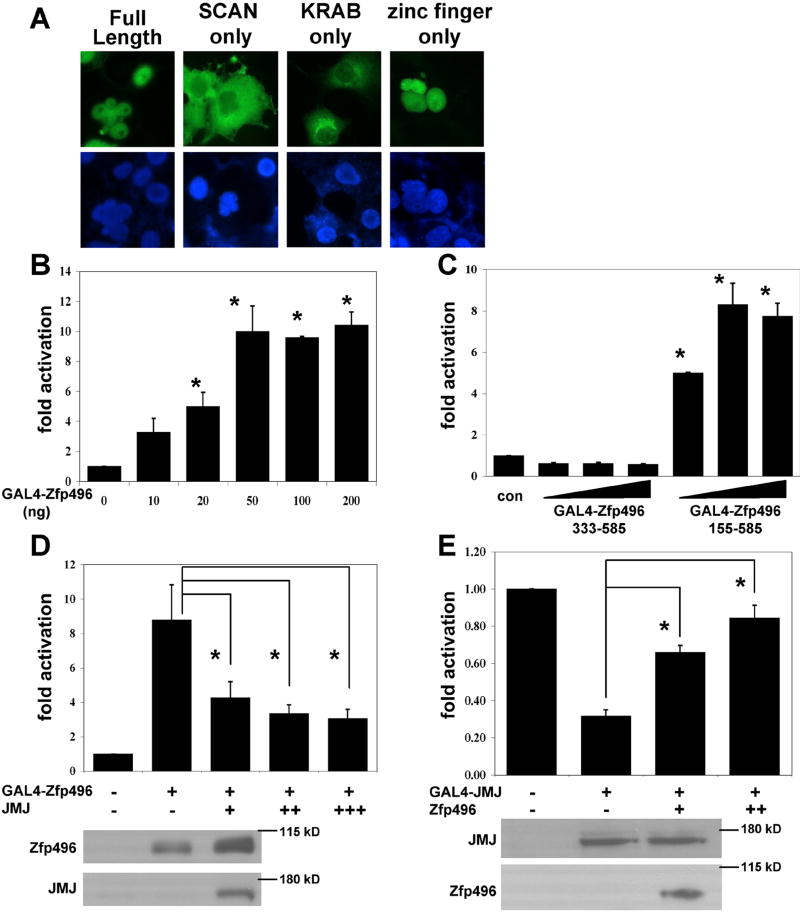
Next, we identified where the nuclear localization signal (NLS) of Zfp496 was located. This information is necessary when examining transcriptional activity of Zfp496 mutants in reporter gene assays. Therefore, deletion mutants of Zfp496 were made in the GAL4-Zfp496-Xpress tag vector. The mutants included the SCAN region alone (aa 1-198), KRAB A box region alone (aa 155-333), and zinc finger region alone (aa 333-585). As Fig. 3A shows, the full length and the zinc finger only mutant localized to the nucleus. Mutants containing only the SCAN region or the KRAB A box region localized to the cytoplasm. Therefore, the NLS is located in the zinc finger region of the Zfp496 protein, which contains a putative NLS sequence, KRRSK (aa 575-579).
Figure 3.
Zfp496 and JMJ modulate each other’s transcriptional activities. A. Determination of the nuclear localization signal (NLS) in Zfp496. COS-7 cells transfected with Zfp496-Xpress mutants were subjected to immunostaining using the anti-Xpress antibody (Invitrogen) and a secondary antibody coupled to AlexaFluor488 (Molecular Probes) (top panel). Nuclei were stained with Hoechst 33342 (Molecular Probes) (bottom panel). Fluorescence was visualized using a Zeiss Axiovert 200 microscope. The structures of the mutants are shown in Fig. 2. B. 10T1/2 cells cultured in 6-well plates were transfected with various plasmids using calcium phosphate. GAL4-Zfp496 was cotransfected with pG5Ti reporter (500ng). C. To map the region of Zfp496 that was responsible for the activation, Zfp496 mutants that localized to the nucleus, GAL4-Zfp496 aa 155-585 (KRAB and zinc finger domain) and GAL4-Zfp496 aa 333-585 (zinc finger only) were cotransfected with pG5Ti (500ng) in varying doses (100ng, 200ng, and 400ng). D. GAL4-Zfp496 (50ng), and pG5Ti (500ng) were cotransfected with or without JMJ (100ng, 200ng, and 400ng). E. GAL4-JMJ (100ng), and pG5Ti (500ng) were cotransfected with or without Zfp496 (20ng and 50ng). Relative luciferase activity was standardized to β-galactosidase activity and the activity was set at one for the reporter gene transfected alone. Each graph represents at least three independent experiments of duplicate plates. Western blots of protein extracts corresponding to first three transfection conditions in D and E are shown.
Based on the fact that Zfp496 contained a KRAB A box and a zinc finger domain, we hypothesized that Zfp496 functions as a transcription factor. A plasmid encoding the GAL4 DNA-binding domain fused to Zfp496 (GAL4-Zfp496) was transfected with a reporter vector pG5Ti consisting of GAL4 DNA-binding sites upstream of the luciferase gene [33]. Interestingly, Zfp496 showed transcriptional activation of the reporter gene up to 10 fold compared to the reporter gene alone in a dose-dependent manner (Fig. 3B). This activation was achieved with a relatively low dose of Zfp496 making it a potent transcriptional activator. This is very interesting because transcription factors containing a KRAB box were generally characterized as transcription repressors. To further characterize Zfp496, we investigated which region of the Zfp496 protein was responsible for the activation by mutational analyses. We cotransfected Zfp496 mutants that localize to the nucleus with the pG5Ti reporter into 10T1/2 cells. The Zfp496 mutant (aa 333-585) containing only the zinc finger region completely lost its activation function compared to that of the reporter gene alone (Fig. 3C). However, Zfp496 (155-585) containing the KRAB box and zinc finger region showed significant activation, in a similar pattern to full length Zfp496. Taken together, these data indicate that the KRAB box and zinc finger region of Zfp496 are required for the activation of the reporter gene.
It is intriguing that Zfp496 shows the transcriptional activation whereas JMJ functions as a powerful transcriptional repressor [24]. We next investigated whether JMJ modulates the activation function of Zfp496 and vice versa since the two proteins showed a physical interaction both in vitro and in vivo. When cells were cotransfected with GAL4-Zfp496 and the reporter pG5Ti, JMJ significantly inhibits the activation function of Zfp496 in a dose-dependent manner (Fig. 3D). JMJ alone did not inhibit the reporter gene activities (data not shown). The reciprocal experiment was performed to determine whether Zfp496 attenuates the repression function of JMJ (Fig. 3E). Zfp496 significantly inhibited the repression function of JMJ in a dose-dependent manner. Zfp496 transfected with only pG5Ti had no effect on the reporter gene activities (data not shown). These data demonstrate that the two factors attenuate each other’s transcriptional function likely via protein-protein interaction. To eliminate the possibility that JMJ inhibited the activation function of Zfp496 by inhibiting the expression of Zfp496, Western blotting was performed. As shown in Fig 3D and E, cotransfection of JMJ or Zfp496 did not inhibit the expression level of Zfp496 or JMJ, respectively.
Characterization of functional interaction between Zfp496 and JMJ
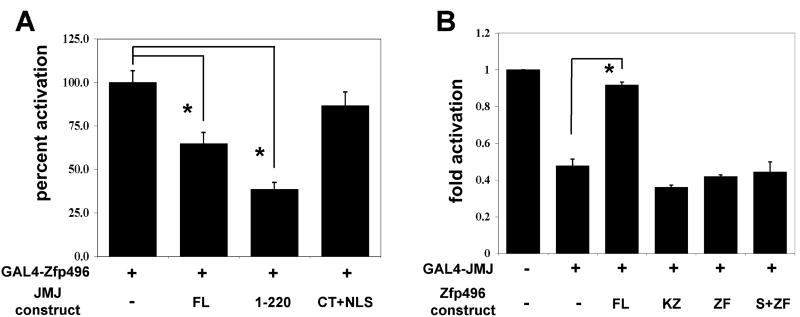
Next we investigated whether the physical interaction between JMJ and Zfp496 is sufficient for this attenuation. GAL4-Zfp496 was transfected with the pG5Ti reporter vector with various JMJ mutants (Fig. 4A). Full length JMJ (FL) and a JMJ mutant containing the NLS and TR domain (1-220) were able to significantly repress the activation of Zfp496. However, a JMJ mutant containing the C-terminal domain fused to the NLS (CT+NLS) showed no significant inhibition of this activation. Because J-Ct (aa 529-1234) localizes to the cytoplasm, the NLS was fused to this mutant to allow the J-Ct to be expressed in the nucleus. These data indicate that the TR domain of JMJ, which is the physical interaction domain, is necessary and sufficient to attenuate the transcriptional activation function of Zfp496.
Figure 4.
The regions of Zfp496 and JMJ responsible for modulating each other’s transcriptional activities. 10T1/2 cells cultured in 12-well plates were transfected with various plasmids using Lipofectamine 2000. A. GAL4-Zfp496 (75ng) was cotransfected with pG5Ti reporter (200ng) and different JMJ constructs. Results are shown where GAL4-Zfp496 was set at 100%. B. GAL4-JMJ (150ng), and pG5Ti (200ng) were cotransfected with different Zfp496 constructs. Relative luciferase activity was standardized and calculated as described in Fig. 3. Each graph represents at least three independent experiments of duplicate plates.
We next investigated whether the zinc finger region, the physical interaction domain, was sufficient for the functional inhibition of the transcriptional repression of JMJ. GAL4-JMJ was transfected with the pG5Ti reporter vector along with various Zfp496 mutants that showed nuclear localization (Fig. 4B). Full length Zfp496 (FL) was able to significantly inhibit the repression of GAL4-JMJ. However, constructs of Zfp496 comprised of KRAB + zinc finger only (KZ), zinc finger only (ZF), and SCAN + zinc finger only (S+ZF) showed no significant inhibition of this repression. These data indicate that while the zinc finger region of Zfp496 is required for a physical interaction with JMJ, it is not sufficient to modulate the transcriptional repression function of JMJ. Rather, all three defined domains of Zfp496 seem to be required for a functional interaction with JMJ. However, we cannot exclude a possibility that deletions in the Zfp496 mutants cause conformational changes in Zfp496 resulting in a loss of functional modulation.
Zfp496 can bind to DNA and activate reporter genes containing its DNA-binding sites
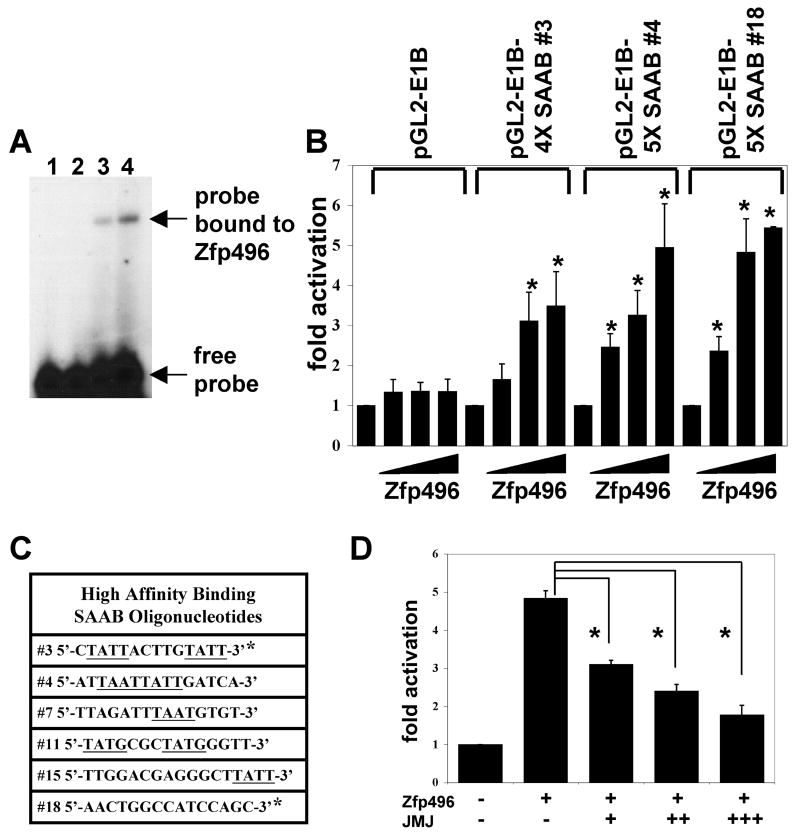
Thus far our data indicates the transcriptional activation function of Zfp496 when it is tethered to DNA using a GAL4 reporter system. Next, we sought to determine if Zfp496 was able to bind to DNA by performing SAAB since the zinc finger domain can function as a DNA-binding domain. Our SAAB assays produced 16 possible binding oligonucleotides in vitro, all of which were confirmed to bind to Zfp496 using GMSA (see Fig. 5A). Eight of the SAAB oligonucleotides showed high affinity binding while the rest of the SAAB oligonucleotides showed lower affinity binding. Of the eight oligonucleotides showing high affinity binding, two oligonucleotides (#3, and #18) were identified twice, suggesting these two oligonucleotides are likely bona fide binding sites for Zfp496 (Fig. 5C).
Figure 5.
Zfp496 can bind DNA and activate SAAB reporter gene constructs. A. Representative GMSA using GST-Zfp496 and the DNA-binding motif #18 selected by SAAB. Lane 1: #18 probe alone. Lane 2: Probe plus 2.5 μg GST only. Lanes 3 and 4: Probe plus 0.5 μg and 1.5 μg GST-Zfp496, respectively. The binding mixture was subjected to non-denatured PAGE and autoradiographed. B. Transient transfection assays were performed in 10T1/2 cells cultured in 6-well plates using the reporter (50ng) and Zfp496 (50ng, 100ng, 200ng) using Lipofectamine 2000. Relative luciferase activity was calculated as in Fig. 3. C. The oligonucleotide sequences of the high affinity DNA-binding sites selected by SAAB are shown. The underlined letters indicate binding sites of homeobox proteins. * indicates the oligonucleotides that were identified twice each. D. Transfection assays were performed in 10T1/2 cells in 12-well plates using the reporter (300ng), Zfp496 (400ng) and JMJ (250ng, 500ng, 1000ng) using Lipofectamine 2000. Each graph in B and C represents at least three independent experiments of duplicate plates.
We reasoned that the high affinity binding motifs that were enriched by SAAB (#3, and #18) would reflect the functional cis-elements in target gene regulation. Therefore, we investigated whether Zfp496 could activate a reporter gene containing these selected binding sites in pGL2-E1B. Transient transfection assays showed that Zfp496 activated all three reporter genes containing the DNA-binding motifs selected by SAAB in a dose-response manner (Fig. 5B). These data demonstrate that Zfp496 can function as a DNA-binding transcriptional activator. It should be noted that five out of a total of six high affinity binding sites contain a -TAAT- like motif (underlined in Fig. 5C) that is a homeobox protein binding site. Initial comparison of all of the SAAB oligonucleotide sequences revealed no obvious consensus binding sequence. JMJ also represses the function of Zfp496 when the reporter gene contains Zfp496 DNA binding sites. When Zfp496 was transfected with pGL2-E1B-5X SAAB #3 or #4 (data not shown) or #18 (Fig. 5D) with or without JMJ, JMJ showed a similar repression function of Zfp496 as in the GAL4 system. JMJ alone did not repress the activity of the reporter gene (data not shown).
DISCUSSION
In this study, we showed that JMJ physically interacted with Zfp496, a SCAN-KRAB-zinc finger protein in vivo and in vitro. The zinc finger region of Zfp496 was required for interaction with the N-terminal part of JMJ containing the transcriptional repression domain [24]. Further, JMJ and Zfp496 attenuated the transcriptional function of each other when they are coexpressed.
It is interesting that Zfp496 functions as a transcriptional activator, although it encodes a KRAB A domain that generally functions as a repressor. It is not known how Zfp496 can function as an activator (Figs. 3, 4 and 5) or repressor [30]. Increasing evidence indicates that one transcription factor can function as an activator or a repressor depending on cofactors and/or promoter context including bifunctional transcription factor YY1 [37]. The nuclear receptors can activate or silence the target gene expression depending on the cofactors [38,39]. Therefore, it is possible that Zfp496 functions as either an activator or repressor depending on cofactors and promoter context. Our SAAB and GMSA results confirm that Zfp496 is able to bind DNA in vitro and therefore function as a DNA-binding transcription modulator. In addition, all reporter constructs tested containing the DNA-binding sites of Zfp496 identified by SAAB were activated by Zfp496 (Fig. 5B). However, we cannot rule out the possibility that the expression of other target genes would be repressed by Zfp496. Further, the possibility exists that the transient transfection system lacks the cofactors necessary to show a transcriptional repressor function of Zfp496. However, our data strongly favor the transcriptional activation function of Zfp496 since a small dose of Zfp496 exhibited activation of two completely different reporter gene systems (Figs. 3 and 5).
The KRAB box is found in about one-third of the human genes encoding Krüppel-type C2H2 zinc fingers (for review see [29]). KRAB-zinc finger proteins require specific cofactors to show transcriptional repression. Namely, they require both KAP-1/TIF-1B and the SET containing protein SETDB1 to function as a repressor [40]. Our finding of a transcriptional activation function of Zfp496 is supported by recent reports that ZNF641 and ZNF480 [41,42] which contain a KRAB and a zinc finger domain, and ZNF445 [43], which contains a SCAN, KRAB and zinc finger domain weakly activate the reporter genes containing AP-1 or serum response element (SRE) sequence. It should be noted that the mechanisms for the weak/marginal activation shown in these articles have not been investigated. Our mutational assays indicated that the KRAB-zinc finger domains were sufficient for the transcriptional activation function while the zinc finger (nuclear localization signal domain) alone completely lost its activation function.
One of three classes of SCAN family members consists of a SCAN-KRAB-zinc finger modular architecture, where the KRAB domain may be complete or more frequently consist of only the A domain, which is the case for Zfp496. Among more than 500 Krüppel-type zinc finger factors encoded in the human genome, about 17 genes contain some variation of the SCAN-KRAB-zinc finger structure similar to Zfp496. Although the functions of these proteins are largely unknown, some interesting members of this subgroup of SCAN family members are ZNF202, and ZNF215. ZNF202 is the hypoalphalipoproteinemia susceptibility gene that encodes a transcriptional repressor that binds to elements found in genes that participate in lipid metabolism [44]. ZNF215 may be associated Beckwith-Wiedemann syndrome, a disorder characterized by growth abnormalities [45]. These examples illustrate that members of the SCAN-KRAB-zinc finger protein family are involved in a diverse set of biological functions.
Although we show that Zfp496 is able to interact with and modulate the function of JMJ, the biological significance of these interactions remains to be elucidated. Since it has been shown that JMJ is involved in regulation of cell cycle and ANF expression in cardiomyocytes during development [25,26,28], it is plausible that Zfp496 regulates these processes by modulating the function of JMJ. Although not cardiac-specific genes, both factors are co-expressed in the developing heart at a higher level than the adult heart by Northern blot analyses (data not shown). The present study provides the important platform to further study functional importance of this modulation as well as to identify the endogenous target genes that are co-regulated by both factors.
Acknowledgments
We thank Tim Ridolfi for his excellent technical assistance, and Drs. Peggy Farnham and Eric N. Olson for providing reporter constructs. This work was supported in part by the NIH grant HL67050 and American Heart Association grant 0030002N to YL. MM was supported in part by American Heart Association predoctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeobox gene Nkx2-5. Genes Dev. 1995;9:1654–66. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 2.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–60. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 3.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–72. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 4.Olson EN, Perry M, Schulz RA. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- 5.Yu YT, Breitbart RE, Smoot LB, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 1992;6:1783–98. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 6.Basson CT, et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–5. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 7.Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. Embo J. 1997;16:5687–96. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, Shioi T, Kasahara H, Jobe SM, Wiese RJ, Markham BE, Izumo S. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol Cell Biol. 1998;18:3120–9. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganga M, Espinoza HM, Cox CJ, Morton L, Hjalt TA, Lee Y, Amendt BA. PITX2 isoform-specific regulation of atrial natriuretic factor expression: synergism and repression with Nkx2.5. J Biol Chem. 2003;278:22437–45. doi: 10.1074/jbc.M210163200. [DOI] [PubMed] [Google Scholar]
- 10.Svensson EC, Tufts RL, Polk CE, Leiden JM. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci U S A. 1999;96:956–61. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Small EM, Krieg PA. Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: Nkx2-5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev Biol. 2003;261:116–31. doi: 10.1016/s0012-1606(03)00306-3. [DOI] [PubMed] [Google Scholar]
- 12.Plageman TF, Jr, Yutzey KE. Differential expression and function of Tbx5 and Tbx20 in cardiac development. J Biol Chem. 2004;279:19026–34. doi: 10.1074/jbc.M314041200. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE. Jumonji, a nuclear protein that is necessary for normal heart development. Circ Res. 2000;86:932–8. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–22. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- 15.Jung J, Mysliwiec MR, Lee Y. Roles of JUMONJI in mouse embryonic development. Dev Dyn. 2005;232:21–32. doi: 10.1002/dvdy.20204. [DOI] [PubMed] [Google Scholar]
- 16.Gregory SL, Kortschak RD, Kalionis B, Saint R. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol Cell Biol. 1996;16:792–9. doi: 10.1128/mcb.16.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheuermann R, Tucker PW. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cellspecific trans-activator that describes a new DNA-binding protein family. Genes Dev. 1995;9:3067–82. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- 18.Kortschak RD, Tucker PW, Saint R. ARID proteins come in from the desert. Trends Biochem Sci. 2000;25:294–9. doi: 10.1016/s0968-0004(00)01597-8. [DOI] [PubMed] [Google Scholar]
- 19.Balciunas D, Ronne H. Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem Sci. 2000;25:274–6. doi: 10.1016/s0968-0004(00)01593-0. [DOI] [PubMed] [Google Scholar]
- 20.Clissold PM, Ponting CP. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem Sci. 2001;26:7–9. doi: 10.1016/s0968-0004(00)01700-x. [DOI] [PubMed] [Google Scholar]
- 21.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 22.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–95. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Gray SG, et al. Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J Biol Chem. 2005;280:28507–18. doi: 10.1074/jbc.M413687200. [DOI] [PubMed] [Google Scholar]
- 24.Kim TG, Kraus JC, Chen J, Lee Y. JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J Biol Chem. 2003;278:42247–55. doi: 10.1074/jbc.M307386200. [DOI] [PubMed] [Google Scholar]
- 25.Kim TG, Chen J, Sadoshima J, Lee Y. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol Cell Biol. 2004;24:10151–60. doi: 10.1128/MCB.24.23.10151-10160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toyoda M, et al. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev Cell. 2003;5:85–97. doi: 10.1016/s1534-5807(03)00189-8. [DOI] [PubMed] [Google Scholar]
- 27.Ohno T, Nakajima K, Kojima M, Toyoda M, Takeuchi T. Modifiers of the jumonji mutation downregulate cyclin D1 expression and cardiac cell proliferation. Biochem Biophys Res Commun. 2004;317:925–9. doi: 10.1016/j.bbrc.2004.03.131. [DOI] [PubMed] [Google Scholar]
- 28.Jung J, Kim TG, Lyons GE, Kim HR, Lee Y. Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J Biol Chem. 2005;280:30916–23. doi: 10.1074/jbc.M414482200. [DOI] [PubMed] [Google Scholar]
- 29.Collins T, Stone JR, Williams AJ. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol. 2001;21:3609–15. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen AL, Jorgensen P, Lerouge T, Cervino M, Chambon P, Losson R. Nizp1, a novel multitype zinc finger protein that interacts with the NSD1 histone lysine methyltransferase through a unique C2HR motif. Mol Cell Biol. 2004;24:5184–96. doi: 10.1128/MCB.24.12.5184-5196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abrink M, Ortiz JA, Mark C, Sanchez C, Looman C, Hellman L, Chambon P, Losson R. Conserved interaction between distinct Kruppel-associated box domains and the transcriptional intermediary factor 1 beta. Proc Natl Acad Sci U S A. 2001;98:1422–6. doi: 10.1073/pnas.041616998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang N, vom Baur E, Garnier JM, Lerouge T, Vonesch JL, Lutz Y, Chambon P, Losson R. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. Embo J. 1998;17:3398–412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fry CJ, Slansky JE, Farnham PJ. Position-dependent transcriptional regulation of the murine dihydrofolate reductase promoter by the E2F transactivation domain. Mol Cell Biol. 1997;17:1966–76. doi: 10.1128/mcb.17.4.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y, Mahdavi V. The D domain of the thyroid hormone receptor alpha 1 specifies positive and negative transcriptional regulation functions. J Biol Chem. 1993;268:2021–8. [PubMed] [Google Scholar]
- 35.Chen CY, Schwartz RJ. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995;270:15628–33. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- 36.Amendt BA, Sutherland LB, Russo AF. Transcriptional antagonism between Hmx1 and Nkx2.5 for a shared DNA-binding site. J Biol Chem. 1999;274:11635–42. doi: 10.1074/jbc.274.17.11635. [DOI] [PubMed] [Google Scholar]
- 37.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 38.Dilworth FJ, Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–54. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- 39.Ordentlich P, Downes M, Evans RM. Corepressors and nuclear hormone receptor function. Curr Top Microbiol Immunol. 2001;254:101–16. doi: 10.1007/978-3-662-10595-5_5. [DOI] [PubMed] [Google Scholar]
- 40.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–32. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi Z, et al. A novel KRAB zinc-finger protein, ZNF480, expresses in human heart and activates transcriptional activities of AP-1 and SRE. Biochem Biophys Res Commun. 2004;320:409–15. doi: 10.1016/j.bbrc.2004.05.182. [DOI] [PubMed] [Google Scholar]
- 42.Qi X, et al. Activation of transcriptional activities of AP-1 and SRE by a new zinc-finger protein ZNF641. Biochem Biophys Res Commun. 2006;339:1155–64. doi: 10.1016/j.bbrc.2005.11.124. [DOI] [PubMed] [Google Scholar]
- 43.Luo K, et al. Activation of transcriptional activities of AP1 and SRE by a novel zinc finger protein ZNF445. Gene. 2006;367:89–100. doi: 10.1016/j.gene.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 44.Wagner S, et al. A broad role for the zinc finger protein ZNF202 in human lipid metabolism. J Biol Chem. 2000;275:15685–90. doi: 10.1074/jbc.M910152199. [DOI] [PubMed] [Google Scholar]
- 45.Alders M, et al. Disruption of a novel imprinted zinc-finger gene, ZNF215, in Beckwith-Wiedemann syndrome. Am J Hum Genet. 2000;66:1473–84. doi: 10.1086/302892. [DOI] [PMC free article] [PubMed] [Google Scholar]