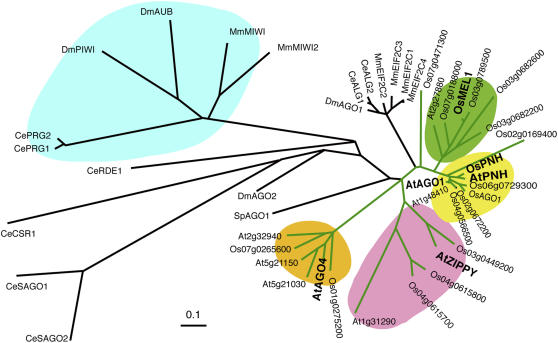
Figure 3.
Phylogenetic Tree Constructed Using the Peptide Sequences of PAZ and PIWI Domains.
For plants, all AGO family members in Oryza sativa (Os) and Arabidopsis thaliana (At) were included (connected with green lines). In addition, functionally known AGOs from Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Mus musculus (Mm), and Schizosaccharomyces pombe (Sp) were included (connected with black lines). The plant locus identifiers correspond to the Arabidopsis Genome Initiative codes for Arabidopsis and the Rice Annotation Project codes for rice. Accession numbers are as follows: NP_510322 (Ce ALG-1), NP_493837 (Ce ALG-2), CAA98113 (Ce PRG-1), AAB37734 (Ce PRG-2), AAF06159 (Ce RDE-1), NP_523734 (Dm AGO1), NP_476875 (Dm PIWI), AAF49619 (Dm AGO2), CAA64320 (Dm AUB), NP_067286 (Mm MIWI), AY135692 (Mm MIWI2), XP_001257025 (Mm EIF2C1), NP_694818 (Mm EIF2C2), NP_700451 (Mm EIF2C3), NP_694817 (Mm EIF2C4), AAD40098 (At PNH), ABO81950 (Os PNH), ABO81951 (Os AGO1), CAA19275 (Sp AGO1), NP_001040938 (Ce CSR-1), NP_504610 (Ce SAGO-1), and NP_490758 (Ce SAGO-2). The PIWI subfamily, which functions specifically in germline development in insects and animals, is highlighted in blue. The plant subfamilies Os MEL1 (AB297928), At AGO1 (AAC18440), At ZIPPY (AAQ92355) and At AGO4 (NP_565633) are highlighted in green, yellow, red, and orange, respectively.