Figure 4.
Map-Based Cloning and Expression Patterns of the HSR8 Gene.
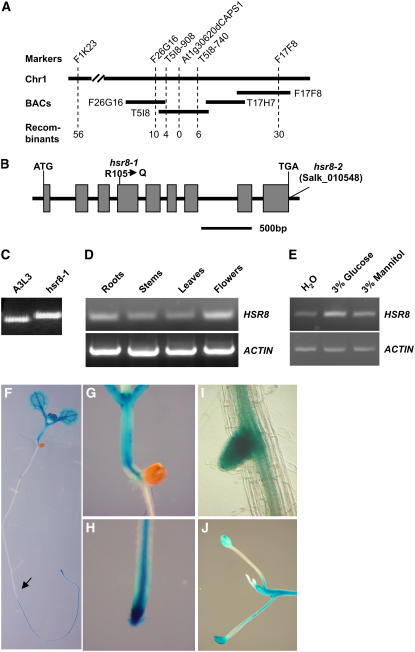
(A) Fine-mapping of the HSR8 locus. The HSR8 locus was mapped to chromosome 1 (Chr1) between markers F1K23 and F17F8. The HSR8 locus was further refined to a 48-kb genomic DNA region between CAPS markers T5I8-908 and T5I8-740 and cosegregated with dCAPS marker At1g30620dCAPS1. The numerals at bottom indicate the number of recombinants identified from F2 plants.
(B) HSR8 gene structure, showing the mutated sites of the two hsr8 alleles. The start codon (ATG) and the stop codon (TGA) are indicated. Closed boxes indicate the coding sequences, and lines between boxes indicate introns. The mutation site in hsr8-1 and the T-DNA insertion site in hsr8-2 also are shown.
(C) The mutation in hsr8-1 was measured with the dCAPS marker At1g30620dCAPS1.
(D) RT-PCR analysis of HSR8/MUR4 gene expression. Total RNA was isolated from roots, stems, leaves, and flowers.
(E) Expression of HSR8 in response to glucose after 6 h of treatment.
(F) to (J) Histochemical analysis of GUS activity in a HSR8pro:GUS seedling (F), hypocotyl and shoot apices (G), a primary root (H), a lateral root (I), and a dark-grown seedling (J).