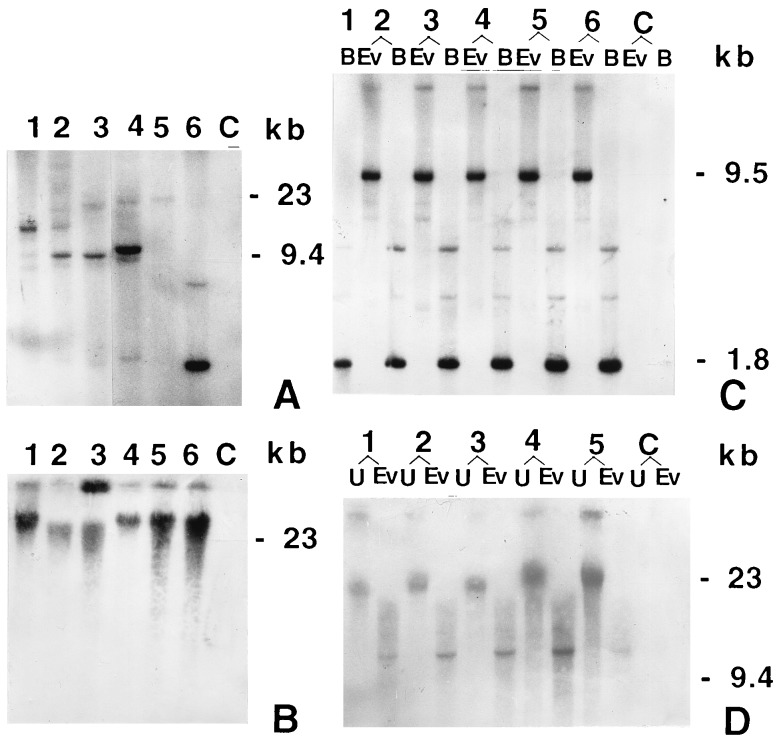
Figure 4.
Southern blot analysis results. The [32P]dCTP-labeled 660-bp DNA fragment from the N-terminal end of the gene was used as the probe for Southern blot analysis. Digested or undigested genomic DNA from each sample was fractionated through an 0.8% agarose gel and blotted to Hybond-N+ nylon membrane (Amersham). After hybridization, the membrane was exposed to Kodak X- Omat film at −70°C. B, BamHI; C, untransformed plant; Ev, EcoRV; U, undigested. (A) Genomic DNA (6 μg per lane) from T0 plants was digested with EcoRV (which has no restriction site in the transforming DNA). Six independent T0 transgenic plants are shown in lanes 1–6, events 3112, 3421, 4017, 756, 1321, and 1804, respectively. (B) Undigested genomic DNA (4 μg per lane) of T0 event 4017 (lane 1) and its five T1 progenies (lanes 2–6). (C) Genomic DNA (6 μg per lane) of the same plants as in B digested with BamHI and with EcoRV. Lanes: 1, T0 event 4017; 2–6, T1 progenies, events 4017-62, 4017-14, 4017-23, 4017-51, and 4017-39, respectively. The EcoRV fragments indicate that event 4017 has two linked integration sites. The fainter high molecular weight band probably represents a single copy, whereas the 9.4-kb band represents multiple copies of the gene in a single site. (D) Genomic DNA (6 μg per lane) of T0 event 3112 (lane 1) and four T1 progenies (lanes 2–5, events 3112-20, 3112-27, 3112-38, and 3112-42, respectively) were digested with EcoRV or not digested. Among the transgenic lines, the presence of a single band in the EcoRV-digested lanes is evident.