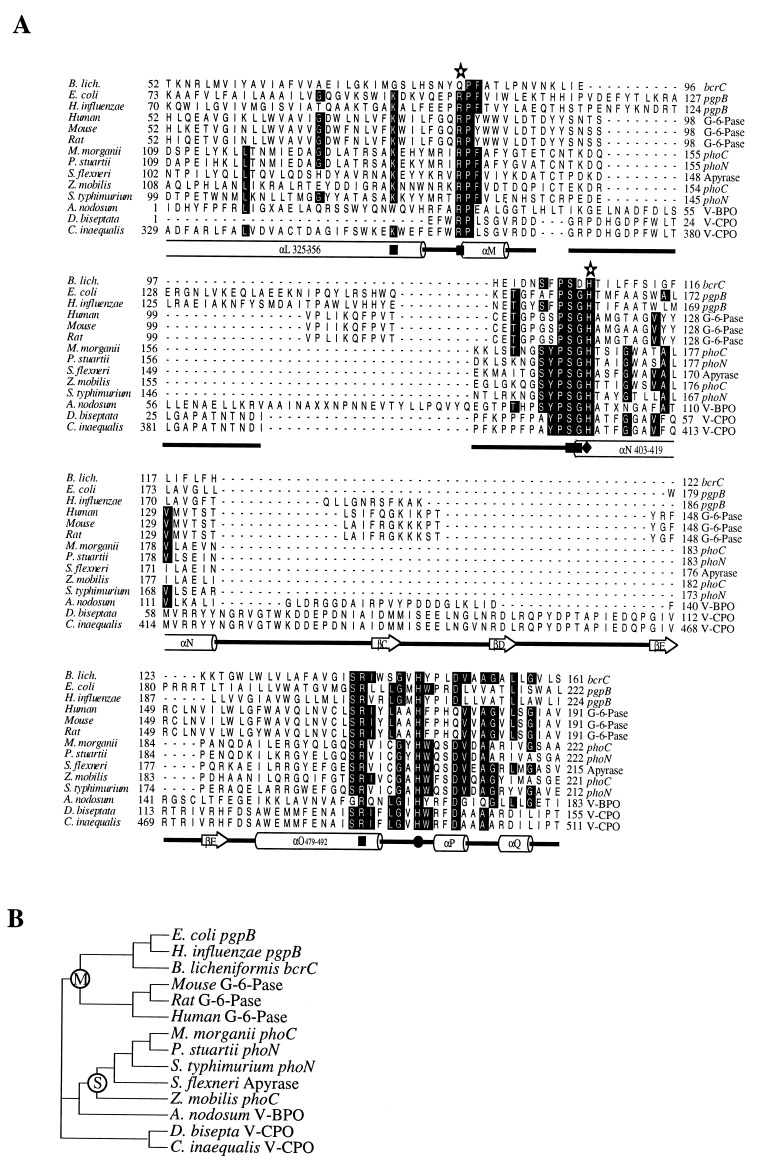
Figure 1.
(A) Alignment of V-CPO of C. inaequalis with the enzymes proposed to contain a structurally similar active site. Residues identical in 50% or more of the sequences are boxed. The residues contributing to the active site of V-CPO are given underneath the alignment together with the secondary structure elements in which they are present. ▪, Residue hydrogen-bonded to vanadate in V-CPO; ♦, histidine proposed to be the acid-base group of V-CPO. •, histidine covalently linked to vanadate in V-CPO; ✫, residue shown to be essential for glucose-6-phosphatase (G-6-Pase) activity; ((), α-helix; ➱, β-sheet; , loop. (B) A dendrogram based on the alignment. The group of membrane-bound phosphatases is marked with an M, and the group of soluble phosphatases is marked with an S. pgpB, phosphatidyl glycerophosphate phosphatase B from Escherichia coli (19) and Haemophillus influenzae (20); bcrC, gene product from Bacillus licheniformis (21); G-6-Pase, glucose-6-phosphatase from mouse (22), rat (23) and human (24); phoC, phoN, class A bacterial acid phosphatase from Morganella morganii (25), Providencia stuartii (GenBank accession no. X64820X64820), Salmonella typhimurium (26), and Zymomonas mobilis (27); Apyrase ATP-diphosphohydrolase from Shigella flexneri (GenBank accession no. U04539U04539); V-BPO partial deduced amino acid sequence from A. nodosum (13); V-CPO partial deduced amino acid sequence from Drechslera biseptata (GenBank accession no. Y11123Y11123) and C. inaequalis (16) (of which the complete sequence is known). The alignment was created after a database search (Swiss-Prot) with small stretches of amino acid residues from the active site of V-CPO of C. inaequalis using the EMBL blitz server. The alignment was created with the seqapp and seqvu programs, and the dendrogram was created using the treeview program (from R. D. M. Page, 1996).