Figure 1.
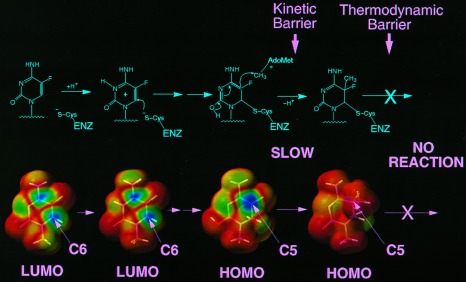
Attack of 5-fluorodeoxycytosine by DNA (cytosine-5)methyltransferases. (Upper) Chemical depiction of intermediates. Protonation of cytosine-N3 by a group at the active site of the enzyme activates the ring for nucleophilic attack by a cysteine residue also at the active site. Once nucleophilic attack occurs, a 5,6 saturated enol can attack the methyl group on S-adenosylmethionine to generate a dihydrocytosine intermediate. In the normal reaction this intermediate would undergo β-elimination to generate a 5-methylcytosine product in DNA and active enzyme. 5-Fluorodeoxycytosine blocks β-elimination because the fluorine at C5 cannot be abstracted. (Lower) Quantum chemical depiction of models of the intermediates. Ab initio geometries were calculated at the Hartree–Fock level of theory using the STO-3G basis set with spartan 4.0 (Wavefunction, Irvine, CA) running on a network of Silicon Graphics workstations. Single-point ab initio orbital energies calculated with the 6-31G* basis set were used to construct electron density surfaces with color maps of frontier orbital values. Blue indicates a high value for the orbital, and red indicates a low value. High values for the lowest unoccupied molecular orbital (LUMO) are seen over C6 and C4 for both 1-methylcytosine and N3 protonated 1-methylcytosine, indicating that these are potential sites for nucleophilic attack. The 1-methyl-6-sulfhydryl-enol derivative of cytosine was used to model the intermediate formed by the enzyme after nucleophilic attack at C6. High values of the highest occupied molecular orbital (HOMO) are confined to C5. This highly reactive orbital is then poised for attack on the methyl of S-adenosylmethionine. Once methyl transfer takes place, the high values of the HOMO remain at C5 but are on the side of the ring (hidden from view) opposite the C5 methyl near the fluorine atom in the model compound (1-methyl-6-sulfhydryl-5-methyldihydrocytosine).
