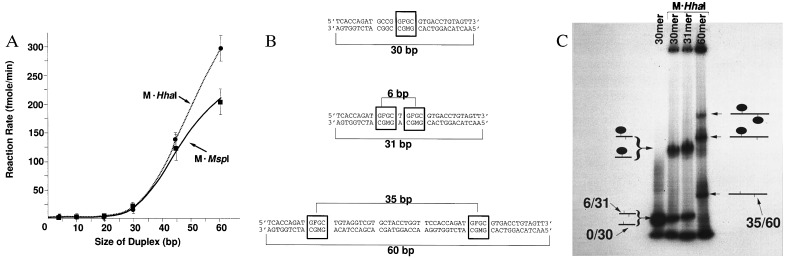
Figure 2.
Spacing in linear molecules. (A) An homologous series of oligodeoxynucleotides each with a single centrally located HhaI recognition site (GCGC/GMGC) was labeled with M·HhaI (2 U) using 4 μM oligodeoxynucleotide at each length. M indicates the position of 5-methylcytosine. These results are compared with data from a second homologous series with the same DNA sequence except that the centrally located tetramer was replaced with the (CCGG/MCGG) sequence recognized by M·MspI. M·MspI (2 U) was used to label 4 μM oligodeoxynucleotide at each length. (B) Three oligodeoxynucleotides were tested for efficiency of M·HhaI assembly. Boxes indicate positions of recognition sites. (C) Nondenaturing gel (using a 6–20%, exponential acrylamide gradient) of the assemblies formed between M·HhaI and the oligodeoxynucleotides depicted in B. 0/30, the 30-mer in B with a single recognition site for M·HhaI (GFGC/GMGC); 6/31, the 31-mer in B with two recognition sites for M·HhaI spaced so that the distance between 5-fluorocytosine residues is 6 bp; 35/60, the 60-mer in B with two recognition sites for M·HhaI spaced so that the distance between 5-fluorocytosine residues is 35 bp. Several minor bands are present in these gels at low concentration. These bands appear to be due to the intrinsic property of oligodeoxynucleotides to form unusual DNA structures, since they are generally present in the absence of added enzyme (e.g., 30-mer lane on the far left). They are not due to impurities in the enzyme preparation because it contained only a single species with a mobility corresponding to the apparent molecular weight of 40,000 kD observed in SDS/gel electrophoresis.