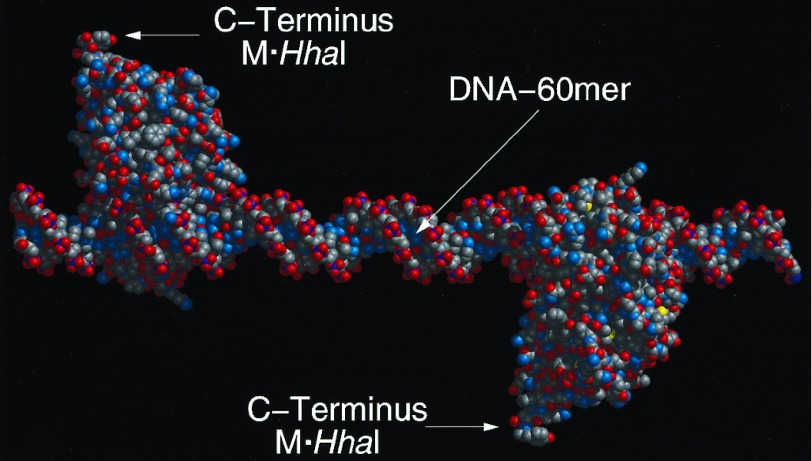
Figure 3.
Molecular model of the 60-mer assembly. Molecular models were constructed in biograf 3.21 (Molecular Simulations, San Diego). The initial conformation of M·HhaI was that determined for the crystalline protein complexed with 5-fluorocytosine at its target site (21). The structure was minimized in molecular mechanics to 0.1 (kcal/mol)/Å, and rendered in midasplus (Computer Graphics Laboratory, University of California, San Francisco). The model assumes a linear DNA molecule with the pitch of 10.0 bp/turn (i.e., a helical twist of 36.0°/bp) derived from fiber diffraction. With this assumption the C termini of the two enzymes assume the 180° dihedral angle, measured down the helical axis, shown in the model. Unwinding of the helix within the DNA binding site has been observed in the M·HhaI DNA complex. Although the DNA is not bent by the enzymes this unwinding could result in a twist angle of 31.6°/bp for those base pairs in the binding site. This consideration, coupled with the possibility that the twist angles for the DNA outside the binding sites could be as low as 34.3°/bp based on solution measurements, suggests that the true dihedral angle could be as low as 80° or as high as the 180° shown in the model above.