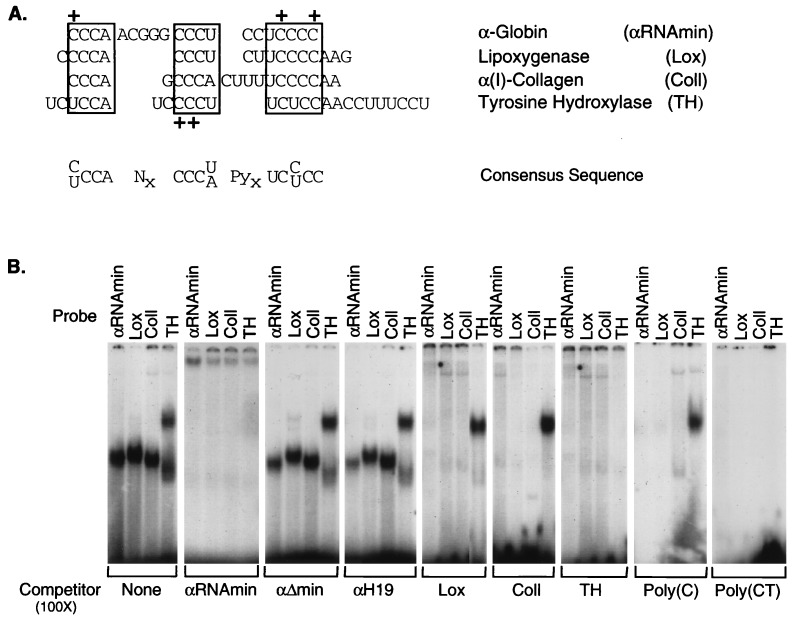
Figure 3.
Structurally related RNP complexes form at pyrimidine-rich segments of 3′UTRs from four stable mRNAs. (A) Sequence alignment of pyrimidine-rich segments within the 3′UTRs of human α-globin (αRNAmin), rabbit 15-lipoxygenase (Lox; ref. 19), human α(I)-collagen (ColI), and rat tyrosine hydroxylase (TH; ref. 18). The sequences shared by all four UTRs are boxed. Gaps are introduced to maximize alignment. A consensus sequence is shown under the sequence alignment. + denotes the positions of mutations within α-globin 3′UTR (ref. 6 and present data) or tyrosine hydroxylase 3′UTR (18) that block RNP complex formation. (B) Gel shift analysis of RNP complex assembly on each of the four 3′UTRs. RNP complexes were assembled and resolved by nondenaturing PAGE as in Fig. 1. The 32P-labeled RNA segment used as a probe in the gel shifts is noted above the lanes, and the unlabeled competitor sequence is noted below each gel. αΔmin and αH19 are two mutant α-3′UTR sequences unable to assemble the α-complex (detailed in text).