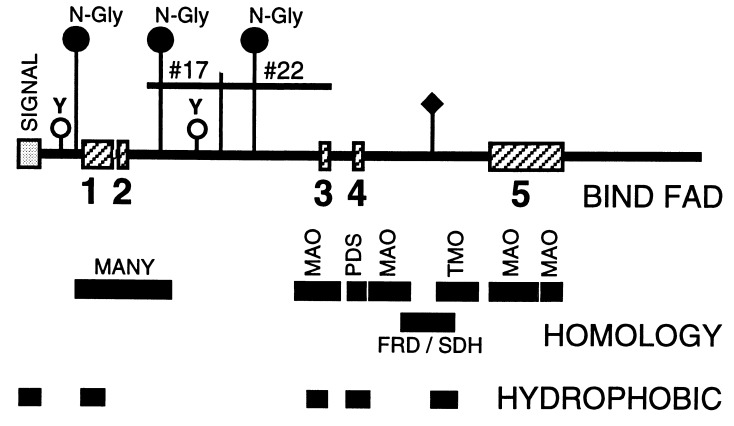
Figure 2.
Schematic of predicted Fig1 protein. The results of computer program-aided sequence analysis of Fig1 is summarized in schematic form. Possible N-linked glycosylation sites (N-Gly) and tyrosine phosphorylation sites (Y) are indicated. Locations of sequence inferred from original subtraction library clones 20-8T#17 and 20-8T#22 are indicated. Predicted hydrophobic regions and FAD cofactor-binding regions (numbered 1–5) are shown. In addition, regions homologous to many known FAD binding and NAD binding proteins (MANY) and to specific FAD binding proteins (MAO, PDS, TMO, and FRD/SDH) are shown. The predicted signal peptide domain is also indicated (SIGNAL). ♦, Location of the homologous FRD/SDH active site. The consensus sequence for the dinucleotide binding fold (FAD-binding region 1) is +-#-X-#-G-X-G-X-X-G-X-X-X-#-X-X-#-X-X-X-X-X-X-#-X-#-X-Δ. +, a hydrophilic, positively charged residue; #, a hydrophobic residue; X, any amino acid; G, glycine; Δ, an acidic residue.