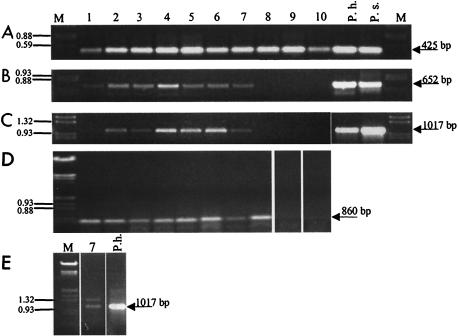
FIG. 2.
Separation of PCR fragments obtained with DNA (A to C) or cDNA (D and E) from 10 sampling sites. The lane numbers indicate the sampling sites (Fig. 1). The following different primer pairs were used on the DNA level: degenerate cyanobacterial 16S rDNA primers CYA359F and CYA781R (A), Prochlorothrix-specific 16S rDNA primers 16S-Pholl-fw and 16S-Pholl-rev (B), and Prochlorothrix-specific pcbC primers pcbCfw and pcbCrev (C). On the cDNA level, degenerate primers optisi-fw, hlwha-l-rev, and pyfadt-rev for pcbB amplification in nested PCR (D) and Prochlorothrix-specific pcbC primers pcbCfw and pcbCrev (E) were used. DNA from cultures of P. hollandica (P.h.) and P. scandica (P.s.) served as positive controls. Lanes M contained a marker (EcoRI/HindIII-digested λ DNA).