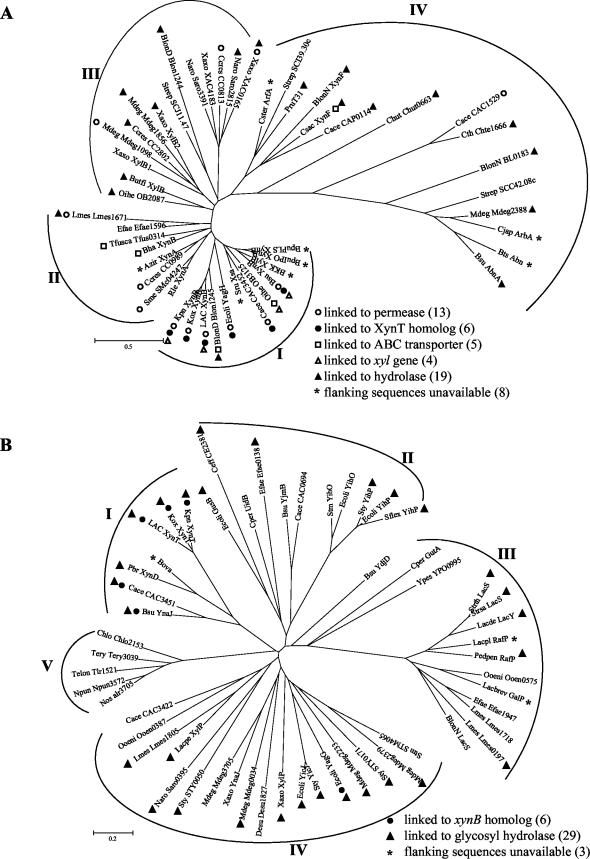
FIG.2.
Unrooted phylogenetic trees of XynB (A) and XynT (B) homologs. Trees have been provisionally assigned into different groups based on similarities in primary structure of XynB (groups I to IV) and XynT (groups I to V), respectively, and the functions of neighboring genes. Abbreviations for the organisms and their XynB and XynT homologs are listed in alphabetical order with accession numbers in parentheses: Azir, Azospirillum irakense, XynA (AAF66622); Bha, Bacillus halodurans, XynB (BAB07402); BKK, Bacillus sp. strain KK-1, XylB (AAC27699); BlonD, B. longum DJO10A, Blon1245 (ZP_00121429), Blon1244 (ZP_00121428); BlonN, B. longum NCC2705, XynF (AAN25335), BL0183 (AAN24037), LacS (NP_696148); BpuPLS, B. pumilus strain PLS, XynB (AAC97375); BpuIPO, B. pumilus strain IPO, XynB (S19729); Bsu, B. subtilis, XynB (AAB41091), AbnA (CAA99586), YnaJ (NP_389639), YjmB (NP_389113), YdjD (NP_388497); Bts, Bacillus thermodenitrificans TS-3, Abn (BAB64339); Butfi, B. fibrisolvens, XylB (A49776); Cace, C. acetobutylicum, CAC3452 (AAK81382), CAC1529 (NP_348156), CAP0115 (NP_149278), CAC3451 (NP_350041), CAC0694 (NP_347331), CAC3422 (NP_350012); Ccres, C. crescentus CB15, CC0989 (AAK22973), CC2802 (AAK24766), CC0813 (AAK22798); Ceff, Corynebacterium efficiens YS-314, CE2381 (NP_738991); Chlo, Chloroflexus aurantiacus, Chlo2153 (ZP_00019154); Chut, Cytophaga hutchinsonii Chut0663 (ZP_00117295); Cjap, C. japonicus, ArbA (CAA71485); Cper, C. perfringens, UidB (NP_561069), GutA (NP_561685); Csac, Caldicellulosiruptor saccharolyticus, XynF (AAB87371); Cster, Caldicellulosiruptor stercorarium, ArfA (CAD48310); Cth, Caldicellulosiruptor thermocellum ATCC 27405, Chte1666 (ZP_00061257); Desu, Desulfitobacterium hafniense, Desu1827 (ZP_00098712); Ecoli, E. coli K12, YagH (P77713), YagG (NP_414804), GusB (AAA68924), YihO (NP_418312), YihP (AAC76874), YicJ (AAC76681); Efae, Enterococcus faecium, Efae1596 (ZP_00036717), Efae0138 (ZP_00035300), Efae1947 (ZP_00037059); Kpn, K. pneumoniae unfinished genome sequence (NC_002941; Washington University, http://genome.wustl.edu); LAC, L. lactis subsp. lactis IL-1403, XynB (AAK05603), XynT (NP_267662); Lacpe, L. pentosus, XylP (P96792); Lacbrev, L. brevis, GalP (AAK54067); Lacpl, L. plantarum, RafP (AAL09166); Lacde, L delbrueckii subsp. bulgaricus, LacY (P22733); Lmes, L. mesenteroides subsp. mesenteroides ATCC 8293, Lmes1671 (ZP_00064177), Lmes1718 (ZP_00064224), Lmes0197 (ZP_00062720), Lmes1805 (ZP_00064310); Mdeg, M. degradans 2-40, Mdeg1098 (ZP_00065724), Mdeg2388 (ZP_00066989), Mdeg1856 (ZP_00066469), Mdeg2233 (ZP_00066837), Mdeg0034 (ZP_00064673), Mdeg3705 (ZP_00068288), Mdeg2379 (ZP_00066980); Naro, Novosphingobium aromaticivorans, Saro3391 (ZP_00096353), Saro2815 (ZP_00095782), Saro0395 (ZP_00093393); Nos, Nostoc sp. strain PCC 7120, alr3705 (NP_487745); Npun, N. punctiforme, Npun3572 (ZP_00109128); Oihe, Oceanobacillus iheyensis, OB3125 (BAC15081), OB2087 (BAC14043); Ooeni, Oenococcus oeni MCW, Ooen0387 (ZP_00069379), Ooen0575 (ZP_00069564); Pbr, P. bryantii, XynD (CAD21012); Pedpen: Pediococcus pentosaceus RafP (P43466); Pru T31, Prevotella ruminicola strain T31 (BAA78558); Rle, Rhizobium leguminosarum bv. trifolii, XynA (AAL14914); Sflex, Shigella flexneri 2a strain 301, YihP (NP_709677); Sme, Sinorhizobium meliloti, SMc04247 (CAC46496); Sru, Selenomonas ruminantium, Xsa (AAB97967); Stm, Salmonella enterica serotype Typhimurium LT2, STM4065 (NP_462946), and YihO (NP_462897); Strep, Streptomyces coelicolor A3 (2), SCJ11.47 (CAB52932), SCC42.08c (CAB92901), and SCI39.30c (CAD55182); Strsa, Streptococcus salivarius, LacS (AAL67293); Strth, S. thermophilus A147, LacS (P23936); Sty, S. enterica subsp. enterica serovar Typhi, STY0171 (NP_454763), YicJ (NP_458178), YihP (NP_458032), and STY0050 (NP_454653); Telon, Thermosynechococcus elongatus BP-1, Tlr1521 (NP_682311); Tery, Trichodesmium erythraeum IMS101, Tery3039 (ZP_00073765); Tfusca, Thermobifida fusca, Tfus0314 (ZP_00056973); Xaxo, Xanthomonas axonopodis pv. citri strain 306, XylB1 (AAM36146), XylB2 (AAM39065), XAC0165 (AAM35057), XAC4183 (AAM39018), XylP (NP_644556), YnaJ (NP_644495); Ypes, Yersinia pestis, YPO0995 (NP_404610).