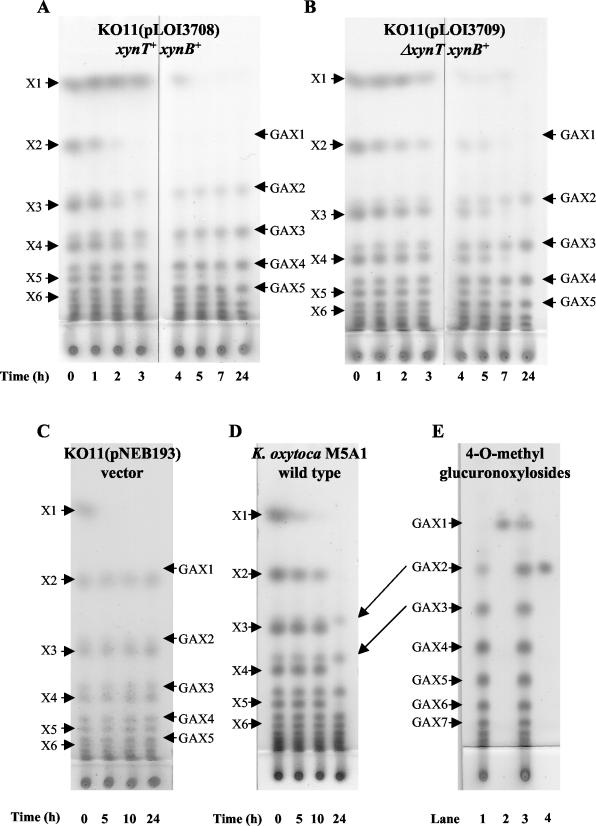
FIG.5.
Thin-layer chromatograms of xylodextrins during fermentation. β-Xylosidase activities for all strains are provided in Table 2. (A) KO11(pLOI3708) containing the full coding regions for xynT and xynB. (B) KO11(pLOI3709) containing the coding region for xynB and a nonfunctional xynT (internal deletion). Note that xylosidase activities in permeabilized cell preparations of KO11(pLOI3708) and KO11(pLOI3709) were equivalent. (C) KO11(pNEB193) containing the vector alone. (D) K. oxytoca M5A1. (E) Cochromatography of GAX1 and GAX2 standards with spent medium from a 24-h xylodextrin fermentation with KO11(pLOI3708). Lanes: 1, spent medium; 2, GAX1 alone; 3, spent medium plus GAX1 and GAX2; 4, GAX2 alone. The labels X1 to X6 refer to the number of xylosyl residues, xylose to xylohexose. Methylglucuronoxylosides containing 1 to 7 xylosyl residues are labeled GAX1 to GAX7, respectively. Arrows connecting panels D and E are included to illustrate positions corresponding to GAX2 and GAX3 in both chromatograms. Xylodextrins were identified by comparison to xylobiose, xylotriose, and cellobiosides as standards (6). Longer methylglucuronoxylosides (GAX3 to GAX7) were inferred by migration rates relative to xylodextrins with equivalent numbers of carbohydrate residues.