TABLE 2.
Homozygous assay
| Genotype | Na | SSA freq.b ± SEM | Pc | GC freq.d | P | N for recute | SSA + GC | N for PCRf | P |
|---|---|---|---|---|---|---|---|---|---|
| A. With [wIw]8z | |||||||||
| Chromosome 2 at 38°g | |||||||||
| +/+ | 30 (1819, 1981) | 0.384 ± 0.029 | 0.415 | Not done | 141 (94) | ||||
| okrWS/Dfh | 26 (2112, 2050) | 0.728 ± 0.045 | <0.0001 | Not done | |||||
| okrRU/Df | 24 (2064, 2110) | 0.822 ± 0.026 | <0.0001 | Not done | |||||
| okrAA/Df | 26 (1946, 1950) | 0.869 ± 0.018 | <0.0001 | Not done | |||||
| Chromosome 3 at 38° | |||||||||
| +/+ | 27 (1445, 1700) | 0.434 ± 0.047 | 0.426 | 411 (366) | 100 (83) | ||||
| spnA1/+ | 28 (1407, 1358) | 0.463 ± 0.043 | 0.6147 | 0.399 | 372 (297) | 100 (95) | |||
| spnA1 | 30 (1965, 1925) | 0.919 ± 0.018 | <0.0001 | Not done | |||||
| spnA1/Dfi | 30 (1647, 1770) | 0.955 ± 0.010 | <0.0001 | <0.001j | 38 (8) | 8 (0) | |||
| B. With [wIw]yellow | |||||||||
| Chromosome X at 38°g | |||||||||
| + | 30 (4739, 4618) | 0.505 ± 0.035 | 0.295 ± 0.024 | 0.800 ± 0.029 | |||||
| lig411 | 29 (4064, 4199) | 0.663 ± 0.047 | 0.0094 | 0.229 ± 0.034 | 0.1177 | 0.892 ± 0.028 | 0.0251 | ||
| Chromosome 2 at 38° | |||||||||
| +/+ | 40 (4424, 4135) | 0.509 ± 0.017 | 0.306 ± 0.017 | 0.815 ± 0.016 | |||||
| mus206A1 | 41 (3809, 3703) | 0.585 ± 0.023 | 0.0093 | 0.295 ± 0.022 | 0.6953 | 0.878 ± 0.013 | 0.0028 | ||
| okrAA/Dfh | 30 (3368, 3956) | 0.868 ± 0.017 | <0.0001 | 0.017 ± 0.007 | <0.0001 | 0.885 ± 0.013 | 0.0016 | ||
| okrRU/Df | 40 (5096, 5181) | 0.829 ± 0.013 | <0.0001 | 0.009 ± 0.004 | <0.0001 | 0.838 ± 0.012 | 0.2453 | ||
| Chromosome 2 at 32° | |||||||||
| +/+ | 40 (3383, 2998) | 0.149 ± 0.017 | 0.306 ± 0.017 | 0.261 ± 0.019 | |||||
| okrAA/Df | 32 (2995, 3098) | 0.543 ± 0.017 | <0.0001 | 0.019 ± 0.006 | <0.0001 | 0.562 ± 0.019 | <0.0001 | ||
| okrRU/Df | 30 (2608, 2470) | 0.431 ± 0.026 | <0.0001 | 0.020 ± 0.008 | <0.0001 | 0.451 ± 0.027 | <0.0001 | ||
N: sample size, the number of male parents tested followed by the number of progeny with and without the I-SceI cut chromosome scored in parentheses.
SSA frequency, the proportion of white-eyed progeny. SEM: standard error of the mean.
The two-tailed P-value calculated using the permutation test for the null hypothesis that the median SSA frequencies are the same between a particular mutant and the WT control. The underlined numbers indicate that the two samples were statistically different.
GC frequencies for the [wIw]8z assay are calculated as follows: 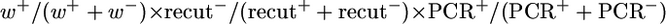 , for testing both the lig4 and spnA mutations with the homozygous assay using the [wIw]8z template. GC frequencies for second chromosome mutations are calculated as follows:
, for testing both the lig4 and spnA mutations with the homozygous assay using the [wIw]8z template. GC frequencies for second chromosome mutations are calculated as follows: 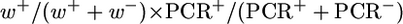 , since w+ progeny were taken for allelic PCR directly without scoring for the recut phenotypes. Since these GC frequencies are derived from complex calculations, no P-values were given for these samples. GC events for the [wIw]yellow assay were scored directly as progeny with light yellow eyes (Figure 2B). GC frequencies are calculated as the proportion of these yellow-eyed flies in total progeny.
, since w+ progeny were taken for allelic PCR directly without scoring for the recut phenotypes. Since these GC frequencies are derived from complex calculations, no P-values were given for these samples. GC events for the [wIw]yellow assay were scored directly as progeny with light yellow eyes (Figure 2B). GC frequencies are calculated as the proportion of these yellow-eyed flies in total progeny.
The number of flies tested for eye mosaicism in the presence of I-SceI to assay the integrity of the I-SceI cut site. The numbers in parentheses are the number of flies producing nonmosaic progeny.
The number of flies tested with allelic PCR to detect interhomolog GC with the [wIw]8z template. The numbers in parentheses are the number of flies producing a positive PCR product.
The heat-shock temperatures for I-SceI induction.
Df=Df(2L)JS17.
Df=Df(3R)X3F.
Since none of the eight w+ recut− events was PCR+, the highest possible GC rate was calculated as 0.001 (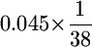 ).
).
