Figure 1.
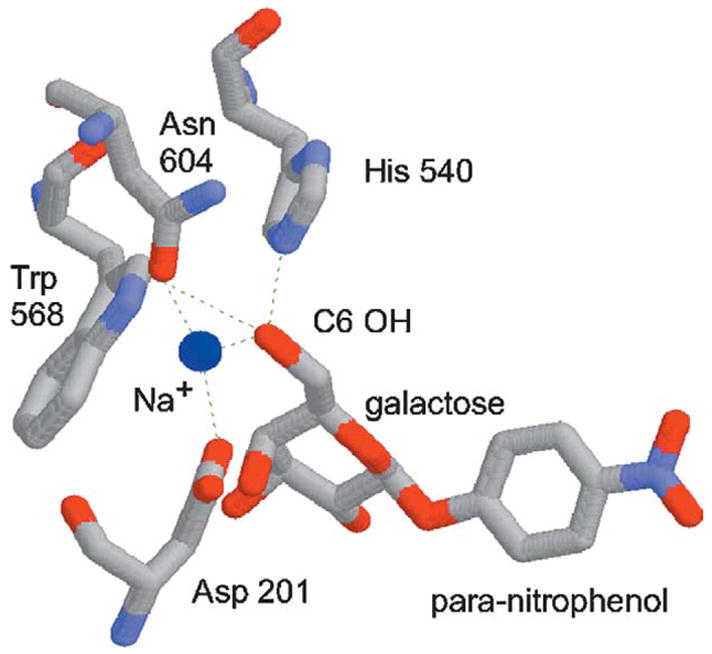
Structure of the Escherichia coli β-galactosidase active site.30 The para-nitrophenyl-β-d-fucopyranoside (“novel substrate”) is identical with para-nitrophenyl-β-d-galactopyranoside (“native substrate”, shown here), except that it lacks the C6 hydroxyl group. The dotted lines represent hydrogen bonds. The Asp201, His540, and Asn604 residues were “randomized” in this study. The sodium ion (blue sphere) is reduced in scale so as not to obscure these amino acid residues.
