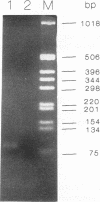
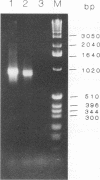
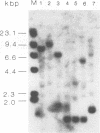
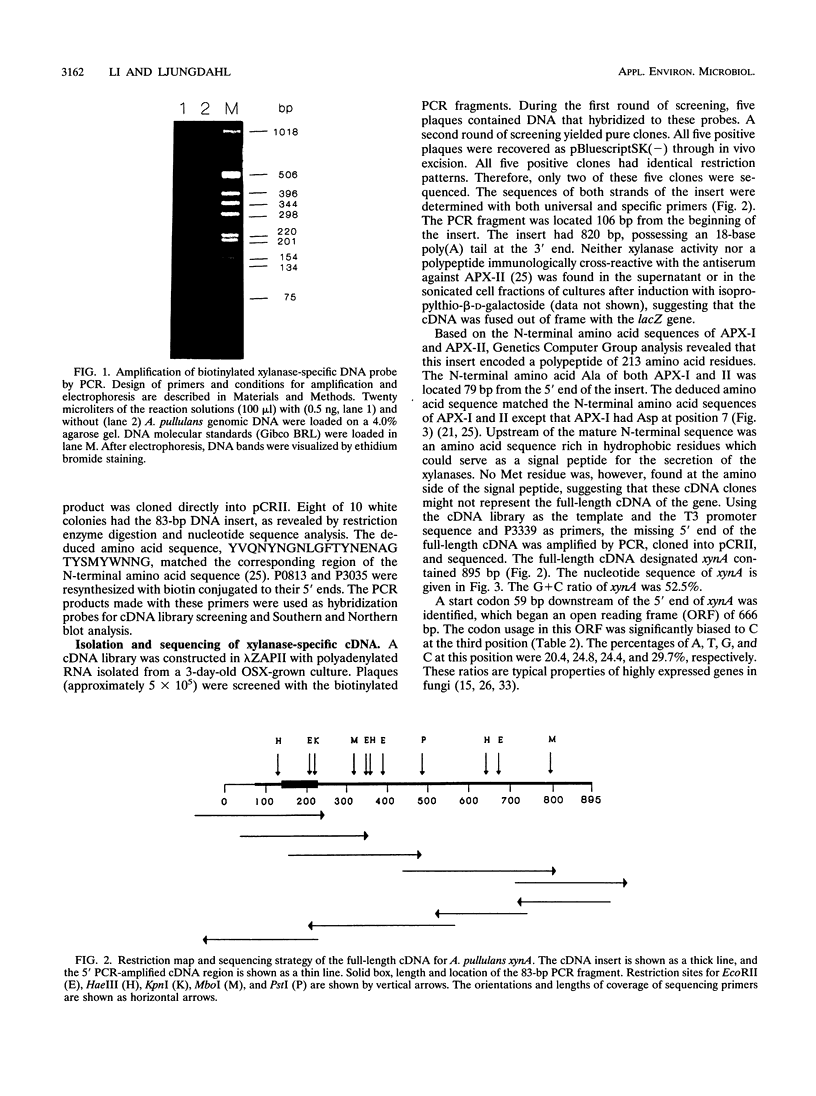
Abstract
Aureobasidium pullulans Y-2311-1 growing on xylan secretes four major xylanases with different masses and isoelectric points. Two of these enzymes, named APX-I and APX-II, have been purified previously. Their N-terminal amino acid sequences are identical except that APX-I has Asp and APX-II has Asn at position 7. An 83-bp DNA region was amplified by PCR and used as a probe for the xylanase gene cloning. The longest cDNA (xynA) obtained by cDNA cloning and PCR amplification consisted of 895 bp. A. pullulans xynA had an open reading frame encoding a polypeptide of 221 amino acids with a calculated mass of 23,531 Da and contained a putative 34-amino-acid signal peptide in front of the amino terminus of the mature enzyme. Strong homology was found between the deduced amino acid sequence of XynA and some xylanases from bacterial and fungal sources. It is suggested that A. pullulans XynA belongs to the family G glycanases. Northern (RNA blot) analysis revealed that only one transcript of 900 bases was present in cultures grown in medium containing D-xylose or oat spelt xylan. Transcription was completely repressed in the presence of glucose in the medium. Southern blot analysis indicated that A. pullulans xynA was present as a single copy in the genome. Comparison between the genomic and cDNA sequences revealed that one intron of 59 bp was present in the coding region. The data presented suggest that the highly active xylanases, APX-I and APX-II, secreted by A. pullulans are encoded by the same gene.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Black M. T., Gunn F. J., Chapman S. K., Reid G. A. Structural basis for the kinetic differences between flavocytochromes b2 from the yeasts Hansenula anomala and Saccharomyces cerevisiae. Biochem J. 1989 Nov 1;263(3):973–976. doi: 10.1042/bj2630973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christov L. P., Prior B. A. Esterases of xylan-degrading microorganisms: production, properties, and significance. Enzyme Microb Technol. 1993 Jun;15(6):460–475. doi: 10.1016/0141-0229(93)90078-g. [DOI] [PubMed] [Google Scholar]
- Gilbert H. J., Hazlewood G. P., Laurie J. I., Orpin C. G., Xue G. P. Homologous catalytic domains in a rumen fungal xylanase: evidence for gene duplication and prokaryotic origin. Mol Microbiol. 1992 Aug;6(15):2065–2072. doi: 10.1111/j.1365-2958.1992.tb01379.x. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B., Claeyssens M., Tomme P., Lemesle L., Mornon J. P. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989 Sep 1;81(1):83–95. doi: 10.1016/0378-1119(89)90339-9. [DOI] [PubMed] [Google Scholar]
- Ito K., Ikemasu T., Ishikawa T. Cloning and sequencing of the xynA gene encoding xylanase A of Aspergillus kawachii. Biosci Biotechnol Biochem. 1992 Jun;56(6):906–912. doi: 10.1271/bbb.56.906. [DOI] [PubMed] [Google Scholar]
- Ko E. P., Akatsuka H., Moriyama H., Shinmyo A., Hata Y., Katsube Y., Urabe I., Okada H. Site-directed mutagenesis at aspartate and glutamate residues of xylanase from Bacillus pumilus. Biochem J. 1992 Nov 15;288(Pt 1):117–121. doi: 10.1042/bj2880117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leathers T. D. Color Variants of Aureobasidium pullulans Overproduce Xylanase with Extremely High Specific Activity. Appl Environ Microbiol. 1986 Nov;52(5):1026–1030. doi: 10.1128/aem.52.5.1026-1030.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. L., Zhang Z. Q., Dean J. F., Eriksson K. E., Ljungdahl L. G. Purification and characterization of a new xylanase (APX-II) from the fungus Aureobasidium pullulans Y-2311-1. Appl Environ Microbiol. 1993 Oct;59(10):3212–3218. doi: 10.1128/aem.59.10.3212-3218.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau A., Durand S., Morosoli R. Secretion of a Cryptococcus albidus xylanase in Saccharomyces cerevisiae. Gene. 1992 Jul 1;116(1):109–113. doi: 10.1016/0378-1119(92)90637-5. [DOI] [PubMed] [Google Scholar]
- Orlean P., Kuranda M. J., Albright C. F. Analysis of glycoproteins from Saccharomyces cerevisiae. Methods Enzymol. 1991;194:682–697. doi: 10.1016/0076-6879(91)94050-m. [DOI] [PubMed] [Google Scholar]
- Ruhnau K., Wegner A. Evidence for direct binding of vinculin to actin filaments. FEBS Lett. 1988 Feb 8;228(1):105–108. doi: 10.1016/0014-5793(88)80595-7. [DOI] [PubMed] [Google Scholar]
- Shareck F., Roy C., Yaguchi M., Morosoli R., Kluepfel D. Sequences of three genes specifying xylanases in Streptomyces lividans. Gene. 1991 Oct 30;107(1):75–82. doi: 10.1016/0378-1119(91)90299-q. [DOI] [PubMed] [Google Scholar]
- Teeri T. T., Lehtovaara P., Kauppinen S., Salovuori I., Knowles J. Homologous domains in Trichoderma reesei cellulolytic enzymes: gene sequence and expression of cellobiohydrolase II. Gene. 1987;51(1):43–52. doi: 10.1016/0378-1119(87)90472-0. [DOI] [PubMed] [Google Scholar]
- Wong K. K., Tan L. U., Saddler J. N. Multiplicity of beta-1,4-xylanase in microorganisms: functions and applications. Microbiol Rev. 1988 Sep;52(3):305–317. doi: 10.1128/mr.52.3.305-317.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Gogary S., Leite A., Crivellaro O., Eveleigh D. E., el-Dorry H. Mechanism by which cellulose triggers cellobiohydrolase I gene expression in Trichoderma reesei. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6138–6141. doi: 10.1073/pnas.86.16.6138. [DOI] [PMC free article] [PubMed] [Google Scholar]