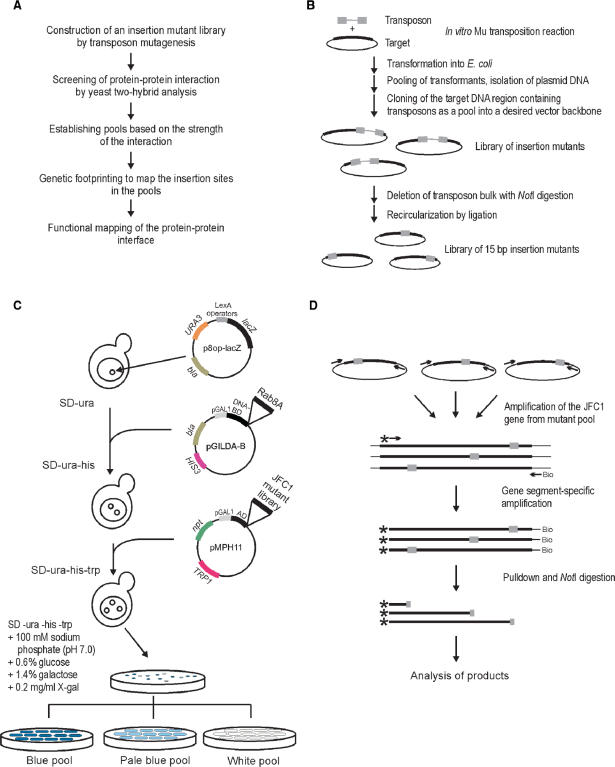
Figure 1.
Experimental outline. (A) Flowchart of the high-precision detection of protein–protein interfaces. (B) Construction of the 15-bp insertion mutant library. (C) Yeast two-hybrid screen. The yeast strain EGY48 carrying the lacZ marker gene-containing plasmid (p8op-lacZ) was transformed with the plasmid (pGildaB-Rab8AΔQ67L) expressing the Rab8A fusion protein and subsequently with the mutant library encoding JFC1 variants cloned in plasmid pMPH11. Plasmid-containing clones were identified on X-gal-containing selection plates. Clones from white, pale blue and blue colonies were collected as streaks and grown to form pools of no, weak or strong protein–protein interaction, respectively. (D) PCR-based genetic footprinting strategy. The JFC1 gene region was first amplified from the insertion mutant library using vector-specific primers. Secondary amplification was done using a biotinylated vector-specific primer and a radioactively labeled JFC1 gene-specific primer. Following pull-down with streptavidin beads, the PCR products were digested with NotI (recognition site within the 15-bp insertion), and the soluble fraction was analyzed by denaturing PAGE and autoradiography.