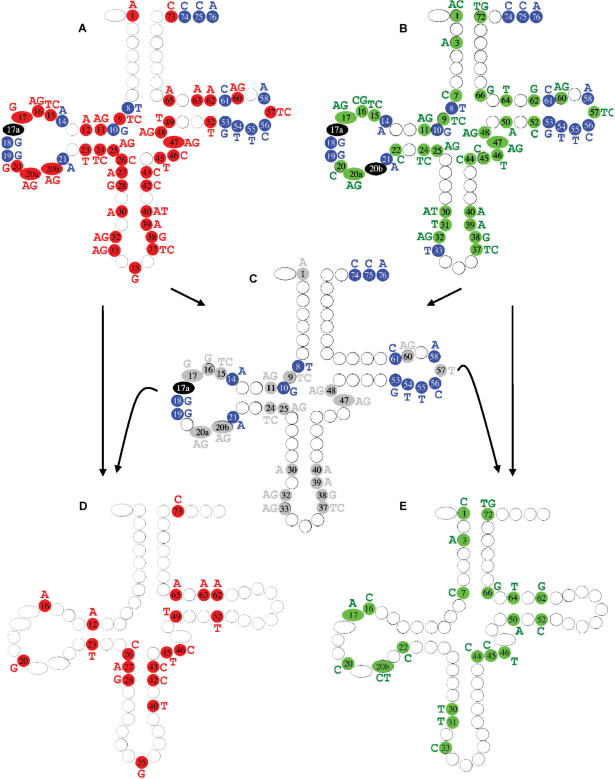
Figure 3.
ECP features of the Saccharomyces cerevisiae tDNA set mapped on the cloverleaf model. The class-specific ECP set for Class I and II are shown in panels A and B, respectively. Strictly present elements are indicated as ‘blue circles’ with white letter, while the strictly absent elements as ‘red circles’ with black letters for Class I, and ‘green circles’ with black letters for Class II. ‘Black circles’ highlight positions where all 4 nucleotide types are strictly absent, corresponding to a gap in the alignment. In panel C we show the intersection of panels A and B, which corresponds to species-specific features characteristic to the entire tDNA set from the given species. ‘Gray background’ indicates the common strictly absent elements of the two classes characteristic to the given species. Note that all of the strictly present elements (blue circles) are species specific, thus no class-specific strictly present elements exist in this species. While the generation of the species-specific strictly absent elements might be self explanatory for most positions, positions like 20 b and 33 might require further explanation. At position 20 b there is a gap in Class II, thus A, C, G and T are all strictly absent elements. Therefore, the intersection with Class I-specific absent elements generates the Class I-specific elements. At position 33 in Class II a T is strictly present meaning that A, C and G are strictly absent. The intersection of the Class I-specific elements, A and G with the Class II-specific elements, A, C and G generates an intersection, A and G. Panels D and E shows the ‘discriminating class-specific elements’ of the strictly absent subset of the ECP for Class I and II, respectively. These sets can be generated as show here: panel D being generated by subtracting panel C from panel A and panel E being generated by subtracting panel C from panel B. Note that the same results are obtained when panel D and E are generated as described in the legend of Figures 1 and 2.