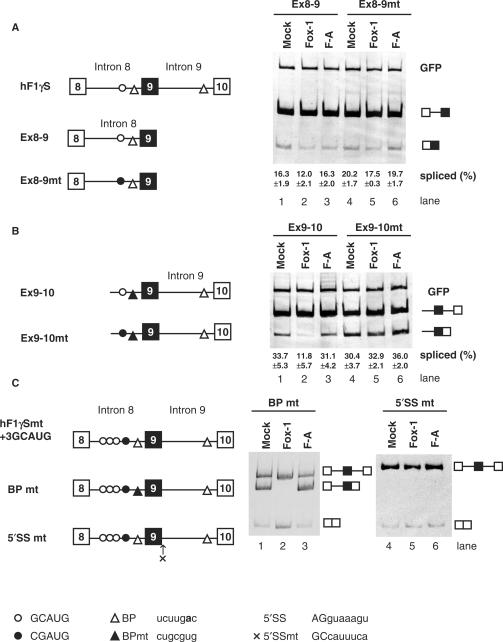
Figure 2.
Mouse Fox-1 induces exon 9 skipping of hF1γ pre-mRNA by repressing the splicing of intron 9 via binding to GCAUG element in intron 8. (A) Analysis of intron 8 splicing in CV-1 cells. The Ex8-9 (lanes 1–3) and Ex8-9 mt mini-genes (lanes 4–6) were co-transfected with pCS2+MT vector (lanes 1 and 4), mFox-1 (lanes 2 and 5), F-A (lanes 3 and 6). The splicing products were analyzed by RT-PCR. A GFP plasmid was cotransfected as an internal reference for transfection efficiency, RNA recovery and loading. The positions of spliced products and GFP are indicated on the right. Average percentage and SD of splicing efficiency are shown at the bottom of each lane. (B) Analysis of intron 9 splicing in CV-1 cells. Transfection analyses of the Ex9-10 (lanes 1–3) and Ex9-10 mt mini-genes (lanes 4–6) with pCS2+MT vector (lanes 1 and 4), mFox-1 (lanes 2 and 5), F-A (lanes 3 and 6). The positions of spliced products and GFP are indicated on the right. Average percentage and SD of splicing efficiency are shown at the bottom of each lane. (C) Transfection analyses of the BPmt (lanes 1–3) and 5′SSmt (lanes 4–6) mini-genes with pCS2+MT vector (lanes 1 and 4), mFox-1 (lanes 2 and 5), F-A (lanes 3 and 6). Schematic representation of mini-genes is shown on the left of each panel. Open and closed circles show the GCAUG element and its mutated element, CGAUG, respectively. Open and closed triangles show branch point (BP) and its mutated site, respectively. A cross represents a mutated 5′ splice site in intron 9. Sequences of these elements are shown at the bottom. A bold letter represents the branch point nucleotide.