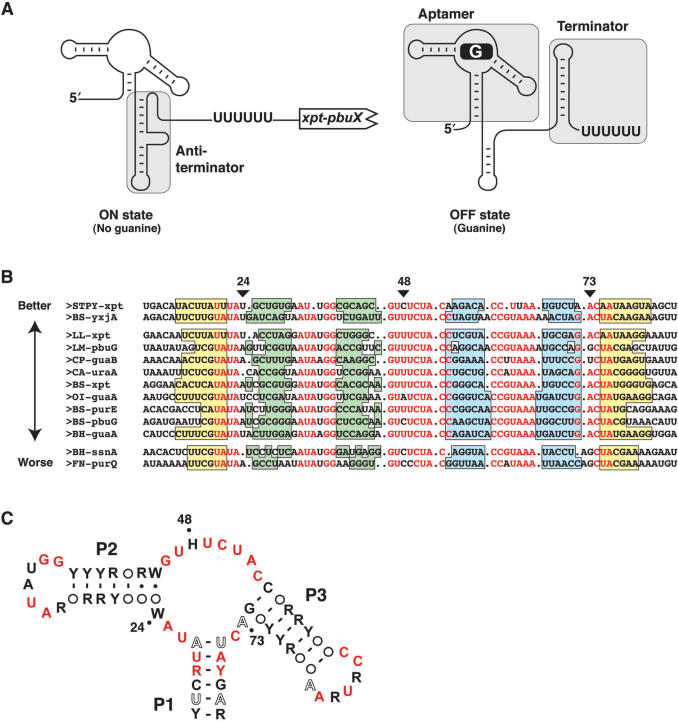
Figure 1.
Consensus sequence of the guanine riboswitch. (A) Schematic representing guanine riboswitch secondary structures in absence (ON state) and in presence (OFF state) of guanine. Shaded regions represent the aptamer, antiterminator and the terminator domains. The polyU sequence known to be important for the transcriptional arrest is shown. (B) Sequence alignment of 13 representative mRNA domains that conform to the guanine riboswitch aptamer motif. Nucleotides in red are conserved in >90% of the sequences. Genes are denoted as previously reported (9). Organisms abbreviations are as follows: Bacillus halodurans (BH), Bacillus subtilis (BS), Clostridium acetobutylicum (CA), Clostridium perfringens (CP), Fusobacterium nucleatum (FN), Lactococcus lactis (LL), Listeria monocytogenes (LM), Oceanobacillus iheyensis (OI) and Streptococcus pyogenes (STPY). Paired regions P1, P2 and P3 are colored in yellow, green and blue, respectively. The three positions of interest in this work are indicated by arrows. Sequences are arranged in three groups corresponding to the 2AP binding activity described in the text. The arrow indicates the relative 2AP binding affinity (Table 1). (C) Consensus sequence and secondary structure of the G box domain. R and Y denote purine and pyrimidine, respectively while H and W indicate A/C/U and A/U, respectively. Circles represent nucleotides not conserved and involved in base pairs. Nucleotides in black, hollow and red represent positions that are conserved >80, >90 and >95%, respectively. The three positions analyzed in this work are indicated by dots.