Abstract
Translational control represents an important mode of regulation of gene expression under stress conditions. We have studied the translation of interferon regulatory factor 2 (IRF2) mRNA, a negative regulator of transcription of interferon-stimulated genes and demonstrated the presence of internal ribosome entry site (IRES) element in the 5′UTR of IRF2 RNA. Various control experiments ruled out the contribution of leaky scanning, cryptic promoter activity or RNA splicing in the internal initiation of IRF2 RNA. It seems IRF2-IRES function is not sensitive to eIF4G cleavage, since its activity was only marginally affected in presence of Coxsackievirus 2A protease. Interferon α treatment did not affect the IRF2-IRES activity or the protein level significantly. Also, in cells treated with tunicamycin [an agent causing endoplasmic reticulum (ER) stress], the IRF2-IRES activity and the protein levels were unaffected, although the cap-dependent translation was severely impaired. Analysis of the cellular protein binding with the IRF2-IRES suggests certain cellular factors, which might influence its function under stress conditions. Interestingly, partial knockdown of PTB protein significantly inhibited the IRF2-IRES function. Taken together, it appears that IRF2 gene expression during stress condition is controlled by the IRES element, which in turn influences the cellular response.
INTRODUCTION
Interferon regulatory factors (IRFs) are DNA-binding proteins that control interferon (IFN) gene expression. IRF1 has been shown to function as an activator of IFN and IFN-inducible genes, whereas IRF2 represses the action of IRF1 (1). Since, interferon induction is followed by translational attenuation; it is plausible that the synthesis of IRF2 protein, which is required to repress and regulate the IFN stimulated genes, is allowed to continue under such condition using an alternate mechanism of translation (2). In fact, the repressor of IFN-β promoter, NRF (NF-κB repressing factor) protein has been shown to be translationally regulated to provide sufficient level of NRF protein for the complete silencing of the IFN-β genes (3).
Initiation of translation is the rate-limiting step of protein synthesis and hence it is tightly regulated. Although the general mode of translation of cellular mRNAs involves cap-dependent translation initiation, a sizeable proportion of mRNAs was shown to be associated with polyribosomes in poliovirus-infected cells at a time when cap-dependent initiation is impaired (4). The most widespread mechanism of cap-independent mode of translation initiation is mediated by internal ribosome entry sites (IRESs), which directly recruits ribosome bypassing the requirement for 5′ cap structure and the cap-binding protein eIF4E (5,6). Many mRNAs that contain IRESs encode proteins that play important roles in cell growth, proliferation, differentiation and regulation of apoptosis (7–10). Stress conditions, such as starvation of growth factors, heat shock, hypoxia and endoplasmic reticulum (ER) stress leads to down regulation of protein synthesis through phosphorylation of eIF2α (11). However, a number of cellular mRNAs containing IRES elements such as vascular endothelial growth factor (VEGF) (12), c-Myc (13), cat-1 mRNA (14), NRF (3) and PITSLRE kinase (15) continue to be translated under conditions when cap-dependent translation is severely impaired. Similarly, inhibition of protein synthesis during apoptosis is accompanied by a caspase-dependent cleavage of initiation factor eIF4G (16). However, there is strong evidence that translation of death associated protein (DAP5) (17), X-chromosome linked inhibitor of apoptosis protein (XIAP) (18), apoptotic protease activating factor (Apaf1) mRNAs is maintained under these conditions and is driven by their respective IRES elements. This indicates that these mRNAs containing IRES may probably have a reduced requirement for the intact eIF4G, allowing the translation of mRNAs containing them to continue under stress conditions (19). This mode of initiation of translation probably protects cells from hostile conditions or at least help them to tide over transient stress conditions.
Here, we have investigated the presence of IRES element in the 5′ untranslated region (UTR) of ‘interferon regulatory factor 2′ or IRF2, which belongs to interferon regulatory factor family (1). Our results suggest that IRF2 5′ UTR (177 nt) contains an IRES element, which undergoes translation initiation in an eIF4G-independent manner. Also, it seems that IFN-α treatment does not inhibit the IRF2-IRES function to the extent observed in case of HCV or BiP IRES activity (GRP78). Analysis of the cellular protein binding with the IRF2-IRES showed specific binding of certain cellular factors, which might influence its function under stress conditions. In fact PTB protein has been shown to specifically interact with the IRF2 5′UTR and partial knock down of PTB protein resulted in significant decrease in IRF2-IRES activity. Additionally, we have studied the effect of ER stress on IRF2-IRES function. In cells treated with tunicamycin, the IRF2 protein level as well as the IRES function was found to be largely unaltered. These results suggest that the IRES element of the IRF2 mRNA allow translation initiation under stress condition and may play a role in the cellular response.
MATERIALS AND METHODS
Plasmid constructs
The cDNA corresponding to the 5′ UTR of IRF2, was amplified from the RNA isolated from HeLa cells and cloned in pCDNA 3.1 (+). The primers were used according to the GenBank sequence NM_002199 and confirmed by DNA sequencing (Gene Bank Acc. No. for IRF2 5′UTR, DQ409328). The construct pRΔENullF was a kind gift from Dr Peter Sarnow (Stanford University). All the bicistronic constructs contain respective 5′UTR sequences (pRIRF2F, pRHAVF and pRBipF) cloned between Renilla luciferase (RLuc) and firefly luciferase (FLuc) genes, in pCDNA 3.1 in between HindIII and EcoRI sites. The eukaryotic promoter less bicistronic construct, the pGEMT-R-IRF-F, containing the IRF2 5′UTR and also the pRΔEnullF bicistronic cassette were cloned in pGEMT easy vector (Promega) under T7 promoter. The T7pRCVB3F was cloned in the pBluescript vector (Stratagene) under T7 promoter. The landscape of structure derived from inactive ΔEMCV IRES sequence was cloned upstream of Rluc gene in the upstream hairpin (uphp) bicistronic plasmid (4). The Nsp bicistronic construct (pRNspF) contains 264 nt from La ORF (120–204 amino acid encoding region) between Rluc and Fluc (20). For constructing IRF2 monocistronic plasmid (pIRF2Fluc), IRF2-Fluc was digested with HindIII and ApaI enzymes (NEB) from the plasmid pRIRF2F and ligated in HindIII, ApaI digested pCDNA 3.1-Fluc. Coxsackievirus 2A protease gene was amplified from CVB3 cDNA (a generous gift from Nora Chapman, Nebraska) using the primers with BamH1 and EcoR1 sites respectively and cloned in pCDN3.1 His C (pCD2Apro). The primers used are as follows: Cox(F):5′ATTAggATCCggCgCATTTggACAA3′; Cox(R):5′ACgCgAATTCCTgTTCCATTgCATC 3′. Bicistronic plasmids pRHAVF and pRBipF were constructed as described earlier (21,22). The primers used for the amplification of IRF2 5′UTR are as follows: IRF2(F)-5′CggCAAgCTTTCTCCTTgTTTTgCT3′;IRF2(R)- 5′ATATgAATTCggTgCCCTCTCAgTg3′.
Cell lines and transfection
Hela S3, Huh7 cells were maintained in DMEM (Invitrogen) with 10% fetal bovine serum (GIBCO, Invitrogen). Cells were transfected with various bicistronic plasmids and pSV40ß-gal using Tfx 20 reagent (Promega) and luciferase assay was performed using Dual luciferase assay reagent (Promega). In experiments using eukaryotic promoter-less bicistronic constructs, cells were infected with vaccinia virus expressing T7 RNA polymerase, VTF7.3 (generous gift from Dr B. Moss, NIH) (23) prior to transfection with bicistronic plasmids. Luciferase assay was performed by dual luciferase assay reporter reagent (Promega) in a TD 20/20 luminometer (Turner Design, CA, USA). For the interferon experiment, Huh 7 cells was transfected with the bicistronic plasmids pRIRF2F, pRHCVF, pRBiPF followed by treatment of 1000 IU/ml of IFN–alpha 2b (Virchow Ltd). For the 2A protease experiment, co-transfection was performed using pRCVB3F, pRIRF2F and pRHAVF bicistronic plasmids with Coxsackie 2Apro plasmid (pCD2Apro) constructs. Luciferase assay was performed after 24 h of transfection. For tunicamycin treatment, cells were incubated in presence of 2.5μg/ml of tunicamycin (Calbiochem) for 14 h. Co-transfection of siRNA with bicistronic plasmid was performed in HeLa S3 cells growing in monolayer using lipofectamine-2000 transfection reagent and optiMEM-I prepared without addition of antibiotic (Invitrogen). Cells were seeded onto 35 mm dishes one day prior to transfection in similar manner. For each transfection, 100 nM of pre-characterized siPTB (Dharmacon) and 1 μg of bicistronic DNA were diluted with optiMEM-I to a final volume of 100 μl. In a separate tube, 6 μl of lipofectamine-2000 was diluted with 94 μl of optiMEM-I to a final volume of 100 μl followed by incubation at room temperature for 5 min. The contents of the two tubes were mixed and incubated at room temperature for 20 min. Subsequently, 800 μl of optiMEM-I was added to the transfection mixture, which was then layered onto cells. Six hours later, the medium was replaced with 2 ml of DMEM (with antibiotic) and 10% FBS. Thirty-six hours post–transfection, the cells were washed, lysed with passive lysis buffer and luciferase enzymes assayed in a similar way. For RNA transfections, capped bicistronic RNAs were synthesized in vitro from different constructs (RIRF2F, RBipF, RΔEnullF) using T7RNA polymerase (Ribomax kit, Promega). Ten microgram of the above synthesized RNAs were used to transfect HeLa cells using Lipofectamine 2000 and optiMEM-I (Invitrogen) as described above. After 6 h, medium was replaced with 2 ml of DMEM (with antibiotic) and luciferase assay was performed by dual luciferase assay reporter reagent (Promega) after 8 h incubation. Enzyme activity was measured in a TD 20/20 luminometer (Turner Design, CA, USA). The transfection efficiency was normalized and the relative luciferase activities were plotted.
In vitro transcription
To make antisense FLuc probe RNA, pCD Luc DNA was linearized with HindIII (NEB) and transcribed by SP6 RNA polymerase (Promega) and 10 μCi/μl of alpha 32P UTP (NEN) as per manufacturer's guidelines. The HCV 5′UTR RNA probe was made from HCV-GFP DNA (22) linearized with EcoRI and was transcribed by T7 RNA polymerase. Similarly, the 32P-labeled RNA probes corresponding to the 5′UTRs of IRF2 and HAV were made from their respective plasmid DNAs after linearizing with either NcoI or EcoRI and transcribed with either Sp6 or T7 RNA polymerase, respectively. The non-specific RNA was made from linearized pGEMT as described elsewhere (22). pRIRF2F, pRBipF, pRΔEnullF bicistronic plasmids were linearized with Pme1 (NEB) and the corresponding bicistronic RNAs were synthesized using Ribomax kit (Promega) following manufacturer's protocol.
Northern blotting and RT-PCR
Total RNA from the HeLa cells, transfected with pRIRF2F, pRCVB3F bicistronic plasmids were extracted using TRIZOL (Sigma), followed by DNase I treatment. Firefly luciferase RNA (Promega) and above extracted RNAs were resolved on a 0.8% agarose–formaldehyde gel, blotted on positively charged Nylon membrane (Millipore) and hybridized with a 32P-labeled riboprobe corresponding to the FLuc gene. Total RNA from HeLa cells transfected with pRIRF2F bicistronic plasmid was extracted using TRIZOL (Sigma). Reverse transcription was performed using AMV RT (Promega) followed by PCR with taq polymerase (Invitrogen).
Western blot hybridization
Huh7 and HeLa cells were harvested and the cell pellet was resuspended in 1× RIPA buffer (10 mM sodium phosphate, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% sodium deoxycholate, 1% NP-40, 0.1% SDS, 0.1% βME, 1 mM PMSF, 50 mM sodium fluoride). Extracts were suspended with 5× SDS gel loading buffer (100 mM Tris-Cl, 200 mM DTT, 4% SDS, 0.2% bromophenol blue, 20% glycerol) and resolved on SDS–10% polyacrylamide gel, followed by electrotransfer of proteins to nitrocellulose membranes. The expression of IRF2 and tubulin was analyzed using antibody specific to IRF2 (a generous gift from Dr Angela Battistini of Istituto Superiore di Sanita, Rome, Italy) and anti rabbit secondary antibody (SIGMA). For detecting tubulin, anti-tubulin antibody (SantaCruz Biotech) was used followed by anti-mouse secondary antibody (SIGMA). TFIID was detected using anti-TFIID antibody (Santa Cruz Biotech) and eIF4G was detected using anti-eIF4G antibody against N-terminal region (Santa Cruz Biotech). The signal was detected by using enhanced chemiluminescence (ECL) detection kit (Amersham-Pharmacia). Similarly for detecting endogenous PTB, anti-PTB antibody was used (Calbiochem).
UV cross-linking
S10 extract was prepared from HeLa and Huh7 cells as described before (21). [α-32P] 5′UTR RNAs were allowed to form complex with S10 extracts as described earlier, followed by cross-linking with UV light. The unbound RNAs were digested with RNaseA treatment. The protein–RNA complexes were then resolved in a SDS–10% polyacrylamide gel followed by phosphorimaging analysis.
Purification of recombinant PTB
The expression of recombinant PTB from PET28a-PTB (a generous gift from Dr J.G. Patton) was induced by 0.6 mM IPTG in Escherichia coli (BL21 DE3) cells transformed with the expression vector. His-tagged protein was purified using Ni2+-nitrilotriacetic acid agarose (Qiagen) under non-denaturing conditions and eluted with 250 mM imidazole.
35S Protein labeling and immunoprecipitation
HeLa cells were treated with either interferon α or tunicamycin for the different time periods as mentioned in the text followed by starvation of the cells for 45 min in MEM-medium lacking methionine (SIGMA). Cells were washed and incubated with 100 μCi of 35S-methionine (trans-label, BARC) for 45 min at 37°C. Cells were harvested and the pellet was resuspended in 2× IP buffer (2% triton X-100 and 0.1% NP40 in TBS) and kept in ice for 1 h. The supernatant was collected and protein estimation was performed using Bradford's reagent (BIORAD). Equal amount of protein was resolved in a SDS–10%PAGE and analyzed by autoradiography. For IRF2 immunoprecipitation, 150 μg of untreated/treated cell extracts were incubated with IRF2 antibody (Santa Cruz Biotech) overnight at 4°C. The immunocomplex was separated by protein A-sepharose beads (SIGMA) for 2 h at 4°C on a rocker. The beads were washed three times with 1× IP buffer and the bound proteins were analyzed by SDS–10% PAGE followed by detection by autoradiogram.
RESULTS
5′UTR of IRF2 is capable of mediating internal initiation of translation
To determine whether IRF2 5′UTR can mediate cap-independent internal initiation of translation to provide basal level of protein under stress; we have investigated the presence of an IRES element in the 5′UTR of IRF2 RNA. For this purpose, IRF2 5′ UTR was amplified by RT-PCR from total RNA isolated from HeLa cells. The nucleotide sequence of the IRF2 5′UTR has been shown in Figure 1A. Zuker's MFOLD algorithm predicted a stable secondary structure with a minimal free energy of −41.7 kcal/mol (Figure 1B) (24). Similar secondary structure was predicted by MFOLD for the BiP IRES (data not shown). It would be interesting to investigate whether IRF2 5′UTR contains the ‘Y’-type stem-loop structure that has been suggested as the characteristic feature of certain cellular IRESs (25).
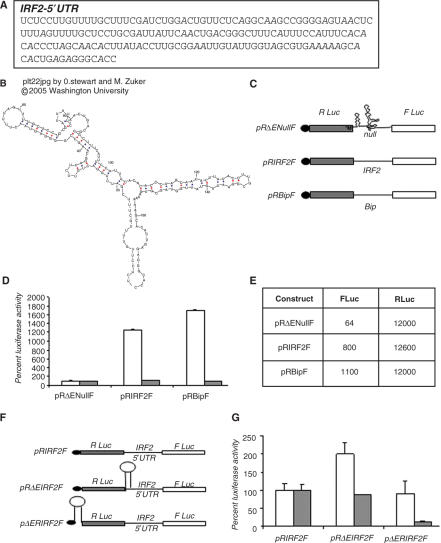
Figure 1.
IRF2 5′UTR sequence mediate internal initiation of translation. (A) Nucleotide sequence of interferon regulatory factor 2 (IRF2) 5′ UTR RNA. (B) MFOLD predicted secondary structure of IRF2 5′UTR. (C) Schematic representation of the bicistronic plasmids used in transient transfections is indicated. (D) Bicistronic plasmids (1 μg) of pRΔEnullF or pRIRF2F or pRBipF were transiently transfected into HeLa cells. Twenty-four hours post-transfection, respective luciferase activities corresponding to Fluc (white bar) and RLuc (gray bar) were measured and shown separately as fold increase compared with that from control (pRΔEnullF) taken as 100%. Transfection efficiencies were normalized by co-transfecting with a β-galactosidase plasmid. The data mean ± SD from three independent experiments. (E) Average absolute values of RLuc and FLuc activities (in relative light units) of the above transient transfection experiments conducted in trplicate are presented in the table. (F) Schematic representation of hairpin containing bicistronic plasmids used in transient transfections is represented. (G) Bicistronic plasmids (1 μg) containing delta EMCV sequence upstream or downstream of RLuc as indicated, were transfected into HeLa cells. The FLuc (white bar) and RLuc (gray bar) activities from the delta EMCV containing plasmids are shown as fold increase or decrease with respect to the corresponding controls, taken as 100. The data mean ± SD from three independent experiments.
To investigate the presence of IRES element in the 5′UTR of IRF2, the amplified IRF2 5′ UTR was subsequently cloned in a bicistronic construct in between two reporter genes. The upstream reporter (Renilla luciferase) in this bicistronic RNA is translated by cap-dependent mode, whereas the downstream reporter (Firefly luciferase) will be translated if the intergenic region contained a functional IRES element. The bicistronic plasmid, pRIRF2F was transiently transfected into HeLa cells followed by luciferase assay. The results showed appreciable amount of firefly luciferase (Fluc) translation mediated by the IRF2 5′UTR. The Fluc activity was found to be around 12.5-fold higher compared to the negative control bicistronic plasmid, pRΔEnullF. Interestingly, the bicistronic plasmid pRBipF, containing Bip IRES as positive control (26), showed ∼17-fold increase in the Fluc activity compared to null bicistronic plasmid control (Figure 1D and E). However, cap-dependent translation of the renilla luciferase (RLuc) was found to be similar in all the three plasmids as expected. These results indicate that the 5′UTR of IRF2 might contain an IRES element, the activity of which is comparable to that of representative cellular IRES.
The cap-independent translation of IRF2 is not due to scanning or ribosomal read-through
In order to rule out scanning or ribosomal read-through as the possible reason for the IRF2 5′UTR-mediated translation of the firefly luciferase, we have used IRF2 bicistronic construct pΔERIRF2F, containing region of highly stable secondary structure upstream of renilla luciferase to prevent ribosome loading (Figure 1F) (4). When the pΔERIRF2F plasmid was transfected into HeLa cells, RLuc activity was found to be significantly inhibited. However, no significant change in Fluc translation was observed compared to values obtained with the control IRF2 bicistronic construct pRIRF2F (Figure 1G). Similarly, when the plasmid pRΔEIRF2F containing the internal hairpin structure inserted downstream of Rluc (Figure 1G) was trasnsfected into HeLa cells, FLuc activity was not inhibited. Interestingly, FLuc activity was found to be marginally higher compared to control, probably due to a change in IRF2-IRES RNA structure in the context of the internal hairpin. However, Rluc activity remained unchanged as expected (Figure 1G). The results suggest that the translation of the downstream cistron Fluc was not due to ribosomal read-through of the first cistron.
IRF2 5′UTR does not have cryptic promoter or splice sites
It is possible that in the cells transfected with IRF2 bicistronic plasmid, small amount of monocistronic FLuc RNA is generated from the bicistronic construct due to cryptic promoter activity of the IRF2 5′UTR sequence. In order to rule out this possibility, HeLa cells were transfected with IRF2 bicistronic construct that was cloned in a vector, which lacked conventional eukaryotic promoter but contained a T7 phage promoter (Figure 2A). Similarly, null and coxsackievirus B3 (CVB3) bicistronic plasmids were used respectively as negative and positive controls in the experiment. Results showed luciferase activity almost equal to background level in absence of any eukaryotic promoter in the transfected constructs. But when cells were transfected with recombinant vaccinia virus expressing T7 RNA polymerase gene (VTF7.3) prior to transfection of the construct, significant levels of Fluc and Rluc activity were detected from the same IRF2 and CVB3 bicistronic constructs (Figure 2A). In fact Fluc activity mediated by IRF2-IRES was found to be around 15 times that of the null construct. As expected, the viral IRES (CVB3) showed much higher efficiency compared to cellular IRES activity under this condition. The result rules out the possibility of cryptic promoter activity in the IRF2 5′UTR.
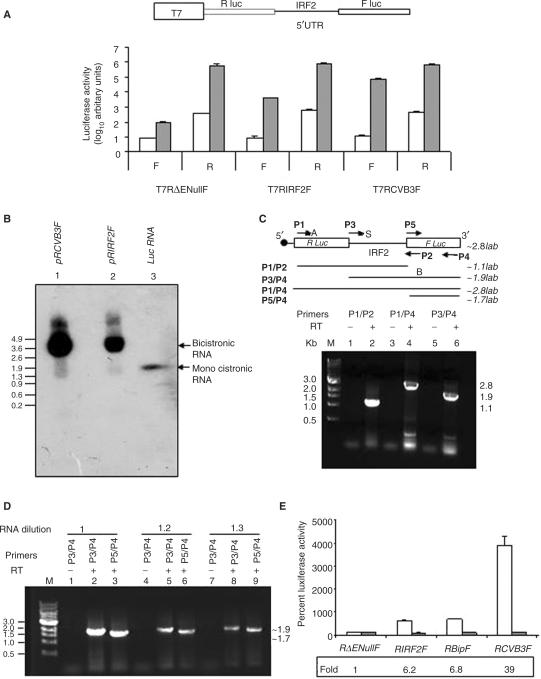
Figure 2.
IRF2 bicistronic plasmid does not show cryptic promoter or splicing activity. (A) Bicistronic plasmid constructs pT7RΔEnullF or pT7RIRF2F or pT7RCVB3F were transfected in HeLa cells in the absence or presence of infection by recombinant vaccinia virus VTF7.3. FLuc activity and RLuc activity were measured and plotted as arbitrary values of log10 separately as indicated. The white bars indicate control (without vaccinia treatment) and the gray bars represent values under vaccinia-treated conditions. The data mean ± SD from three independent experiments. Schematic representation of bicistronic plasmid used in transient transfections is shown above the panel. (B) Northern blot of total RNA extracted from Hela cells transfected with pRCVB3F (lane1), pRIRF2F (lane 2) bicistronic DNAs and FLuc RNA (Promega) (lane 3) using 32P-labeled riboprobe corresponding to FLuc. (C) RT-PCR analysis, using combination of four primers P1/P2 and P3/P4 and P1/P4 (as indicated above the panel) of RNA extracted from pRIRF2F bicistronic plasmid transfected HeLa cells. Lane M is the 1 kb DNA molecular marker. Lanes 2, 4 and 6 depict the amplified product obtained when P1/P2, P1/P4, P3/P4 combination primers were used. Lanes 1, 3 and 5 show reverse transcriptase-negative control for each set of primers. (D) RT-PCR analysis as above, using different dilution of input RNA with different combination of primers as indicated above the lanes. (E) RNA transfections: capped bicistronic RNAs (10 μg) corresponding to RΔEnullF or RIRF2F or RBipF or RCVB3F were transiently transfected into HeLa cells. Relative luciferase activities corresponding to Fluc (white bar) and RLuc (gray bar) were measured and shown separately as fold increase compared to that of negative control (RΔEnullF) taken as 100%. The values below the panel indicate the fold increase in respective IRES activity. The data mean ± SD from three independent experiments.
To rule out the possibility, that IRF2 5′UTR might contain splice sites which generate monocistronic Fluc RNA in vivo, northern blot hybridization assay was performed. For this purpose, total RNA was extracted from the cells transfected with IRF2 bicistronic plasmid and probed with a 32P-labeled riboprobe complimentary to Fluc gene. As a positive control, RNA extracted from cells transfected with a similar bicistronic plasmid containing Coxsackievirus B3 (CVB3) IRES was included in the assay. In our northern blot assay, we failed to detect any smaller RNA products derived from either of the transfected bicistronic plasmids (Figure 2B).
We have also performed RT-PCR analysis from the total RNA extracted from cells transfected with the IRF2 bicistronic plasmid using different sets of primers as shown in Figure 2C, which showed the presence of intact bicistronic RNA in vivo. Additionally, to investigate the presence of shorter monocistronic RNA (if any), we have performed RT-PCR analysis of total RNA as above with different dilutions of input RNA and primer sets as indicated above the panel in Figure 2D. In this experiment primer set P5/P4 would amplify the region of Fluc gene only, whereas the primer set P3/P4 would amplify the full-length IRF2 5′UTR along with the Fluc gene. In the event of cryptic splice sites within IRF2 5′UTR in the bicistronic construct, the ratio of the amplified products generated by using P5/P4 primers would be significantly more than that of P3/P4 product. However, in our assay we did not find significant differences in the amplified products (Figure 2D).
To further validate the IRF2-IRES activity, we have performed RNA transfection experiment. Capped bicistronic RNAs, synthesized in vitro from different plasmid constructs (RΔEnullF RIRF2F, RBipF, RCVB3F) were transiently transfected into HeLa cells and the relative luciferase activities were measured 8h post-transfection. The Fluc activity mediated by IRF2-IRES was found to be 6-fold higher than that obtained for the negative control pRΔEnullF (Figure 2E). Similar fold increase in Fluc activity was observed for Bip IRES, used as positive control for cellular IRES (Figure 2E). Interestingly, the Fluc activity mediated by the viral IRES (CVB3) was found to be much higher than the cellular IRESs. However, the results are consistent with recent reports, which suggest that cellular IRESs are not as active as viral IRESs when RNA transfections are performed, possibly because cellular IRESs require ‘nuclear history’ for their optimum activity (27,28).
Cellular protein binding with the IRF2 5′UTR
To further characterize the IRES activity of IRF2 RNA, we have studied the requirement of canonical and non-canonical initiation factors. Picornavirus 2A protease has been shown to cleave eIF4G and shut down cap-dependent translation of cellular mRNAs (29–31). Although viral IRES elements are not sensitive to eIF4G cleavage, hepatitis A virus IRES has been shown to require intact eIF4G for efficient translation (32). To investigate whether the IRF2-IRES needs intact eIF4G for its efficient function, we have used the Coxsackievirus B3-2A protease encoding plasmid CVB3-2Apro. This plasmid was co-transfected with IRF2 bicistronic plasmid pRIRF2F in HeLa cells. In addition, bicistronic plasmids containing the IRES element of either Coxsackievirus B3 (CVB3) or Hepatitis A virus (HAV) were also included as controls (Figure 3A). Western blot analysis of the CVB3-2A protease-treated cell extract showed cleavage of eIF4G in the transfected cells in vivo (Figure 3B). The expression of CVB3-2A protease resulted in drastic decrease of cap-dependent translation of RLuc in case of all three bicistronic plasmids but did not significantly affect Fluc translation mediated by IRF2 and CVB3 IRESs (Figure 3A). On the other hand, HAV IRES activity was reduced by CVB3-2Apro expression. Thus it appears that the IRF2-IRES-mediated translation might not require the presence of intact eIF4G.
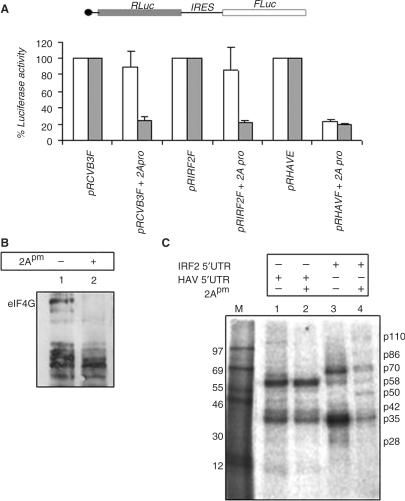
Figure 3.
Effect of CVB3-2Apro on IRF2-IRES activity. (A) 50 ng of pCD2Apro plasmid was transiently co-transfected with 1 μg of pRCVB3F or pRIRF2F or pRHAVF plasmids in HeLa cells. Luciferase activity was measured after 24 h. FLuc (white bar) and RLuc (gray bar) activities are shown as fold decrease with respect to corresponding controls (without 2A pro treatment), taken as 100%. The data mean ± SD from three independent experiments. The absolute values of the firefly luciferase (Fluc) in the absence/presence of 2A protease is as follows: pRCVB3F (1400/1100), pRIRF2F (720/600) and pRHAVF (560/140). (B) 100 μg of cell extract from either HeLa cells transfected with plasmid CVB3-2Apro or untransfected HeLa cells (as control) were resolved in SDS–8% PAGE and probed with anti eIF4G antibody (against N-terminal). Lane 1 corresponds to control HeLa cell extract and lane 2 depicts the 2A protease expressed cell extract showing cleavage of eIF4G. (C) UV cross-linking assay with 10 μg of either mock or 2Apro-treated HeLa S10 extracts (as indicated above lanes) with 32P-labeled HAV 5′UTR (lanes 1 and 2) and IRF2-5′UTR (lanes 3 and 4) RNA probes.
To investigate the cellular protein binding with IRF2 5′UTR, UV cross-linking experiment was performed using HeLa S10. The results showed major binding with p70, p58 and p35 and minor binding with p110, p86 and p42 with the IRF2 5′UTR (Figure 3C). Interestingly, when S10 was used from HeLa cells treated with CVB3-2A protease, a prominent band of 50 kDa polypeptide (p50) was found to cross-link with the IRF2-IRES. However, the same protein did not show interaction with HAV 5′UTR. Also another polypeptide of ∼110 kDa did not show binding with the HAV 5′UTR when 2A protease-treated cell extract was used (Figure 3B, compare lanes 1 and 2). It has been shown earlier that the ∼58 kDa band interacting strongly with the HAV 5′UTR is actually polypyrimidine-tract-binding protein (PTB) (21). To investigate whether PTB protein also interacts with IRF2, UV cross-linking experiment was performed with purified recombinant PTB protein (Figure 4A, lane 1). As expected, 100-and 500-fold molar excess of the cold self IRF2 5′UTR RNA competed out the PTB binding with the radiolabeled 5′UTR probe (Figure 4A, lanes 2 and 3), whereas competition with 100- and 500-fold molar excess of non-specific RNA failed to compete the PTB binding with the IRF2 5′UTR suggesting the specificity of the interaction (Figure 4A, lanes 4 and 5).
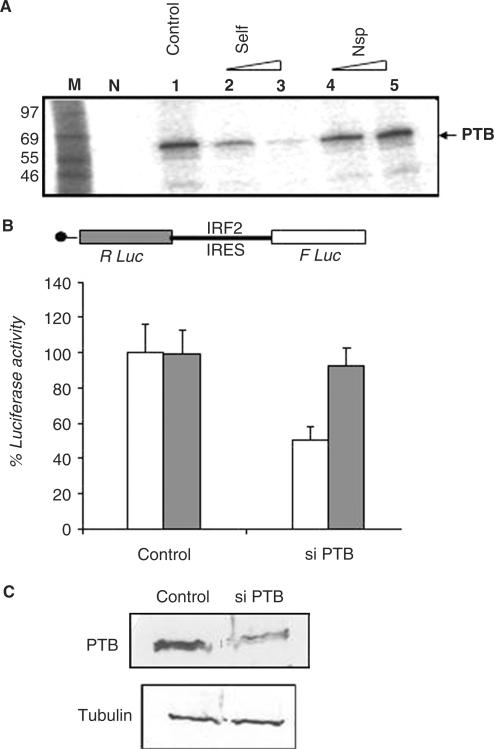
Figure 4.
Binding of PTB with the IRF2-IRES and effect of its silencing on the activity. (A) Purified recombinant PTB protein was incubated with 32P IRF2 5′UTR in absence (lane 1) or presence of 100-(lane 2) and 500-fold (lane 3) molar excess of self-cold IRF2 5′UTR (Lanes 2–3) or non-specific RNA (lanes 4–5). The RNA protein complexes were UV cross-linked and analyzed on SDS 10% PAGE followed by phosphorimaging. Lane M represents the molecular weight marker and lane N represents no protein control. (B) HeLa monolayer cells were transiently transfected with bicistronic plasmids pRIRF2F in absence or presence of 100 nM siPTB. Luciferase activity was measured 36 h post-transfection. FLuc (white bar) and RLuc (gray bar) activities are shown as fold decrease with respect to corresponding control (without siRNA), taken as 100%. The data mean ± SD from three independent experiments. (C) Western blot analysis of the above transfected cell lysates (40 μg), using anti-PTB antibody and anti α-tubulin antibody.
Although the precise mechanism of the cellular IRESs is not clear, the requirements of some auxiliary factors, called as IRES trans-acting factors (ITAFs) is well documented (33). To elucidate the possible role of PTB in modulating the IRES activity of IRF2, we partially silenced PTB by transient gene silencing method using siRNAs. For this purpose, bicistronic plasmid DNA containing IRF2-IRES was co-transfected with 100 nM of a pre-characterized siRNA specific for silencing PTB gene (34). The results showed significant decrease in IRF2-IRES-mediated translation of Fluc whereas cap-dependent translation of RLuc was not affected significantly (Figure 4B). However, a non-specific siRNA did not inhibit IRF2-IRES activity (data not shown). As expected, the western blot analysis showed almost 50% decrease in the PTB protein level in the siPTB-treated cells compared to control (Figure 4C), suggesting that the level of PTB could be critical determinant for the efficient function of the IRF2-IRES element.
Effect of interferon α treatment on the IRF2-IRES activity
IRF2 belongs to the interferon regulatory factor family and is known to have a transcriptional repressor activity (35). We were interested to explore the physiological significance of the IRF2-IRES activity (if any) in regulating cellular response to interferon. Interferon has been shown to inhibit the IRES activity of hepatitis C virus (HCV) (36), and is also known to activate PKR which leads to phosphorylation of eIF2α resulting in inhibition of cap-dependent translation (2).
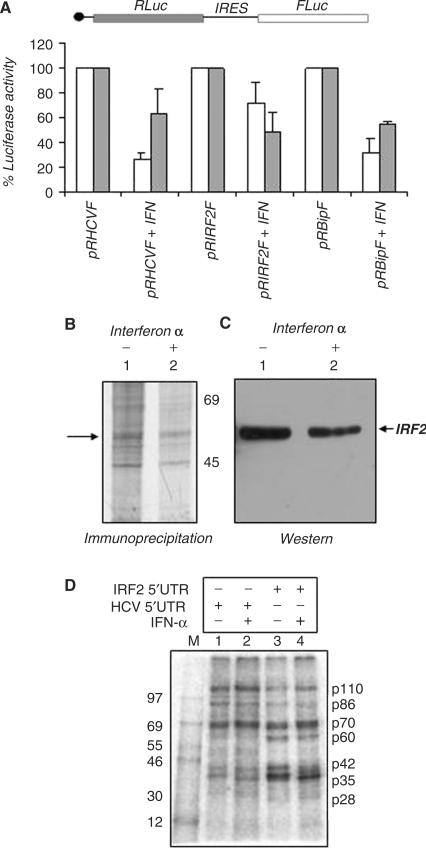
As a first step, we have investigated the effect of interferon treatment on the activity of IRF2-IRES and used HCV IRES or Bip IRES as representative controls. For this experiment, the hepatocellular carcinoma cells (Huh7) was chosen as the experimental cell line, since earlier studies with this cell line have demonstrated the inhibitory effect of IFN α on HCV IRES activity (36). When Huh7 cells were transfected with either HCV or IRF2 or Bip IRES containing bicistronic plasmids and treated with 1000 IU/ml of IFNα for 24 h, it was observed that HCV IRES activity was significantly inhibited as reported earlier (36). However, the extent of inhibition was much less in the case of IRF2-IRES activity as compared to HCV and BiP IRESs (Figure 5A).
Figure 5.
Effect of interferon α (IFN α) treatment on the IRF2-IRES activity. (A) Huh 7 cells were transiently transfected with bicistronic plasmids pRHCVR or pRIRF2F or pRBipF. Twelve hours post-transfection, IFNα (Virchow Ltd) was added at a concentration of 1000 IU/ml. Luciferase activity was measured 24 h post-transfection. FLuc (white bar) and RLuc (gray bar) activities are shown as fold decrease with respect to corresponding control (without IFN treatment), taken as 100%. The data mean ± SD from three independent experiments. The absolute values of the firefly luciferase (Fluc) in the absence/presence of IFN treatment is as follows: pRHCVF (660/160), pRIRF2F (2600/1600) and pRBiPF (1500/480). (B) Immunoprecipitation followed by western of IRF2 in untreated and IFN-treated cell extracts. Cells were metabolically labeled in the absence and presence of interferon (as indicated above lanes). Lanes 1 and 2 correspond to immunoprecipitation of IRF2 using cell extract in absence and presence of IFNα respectively, the arrow indicates the IRF2 protein band. (C) Lanes 1 and 2 correspond to the western blot of IRF2 with the above immunoprecipitated extracts. (D) UV cross-linking assay with 8 μg of either mock or IFNα-treated Huh7 S10 extracts (as indicated above lanes) with 32P-labeled HCV 5′UTR (lanes 1 and 2) and IRF2-5′UTR (lanes 3 and 4) RNA probes.
Additionally, to monitor the synthesis of IRF2 protein during interferon α treatment, pulse metabolic labeling experiment was performed. For this purpose, Huh7 monolayer cells were incubated with interferon α for 12 h, followed by in vivo pulse labeling of proteins using 35S-methionine. The results showed appreciable decrease in overall protein synthesis, although some of the proteins were found to continue synthesis even after interferon α treatment (data not shown). Immunoprecipitation of the pulse-labeled proteins using anti IRF2 antibody showed no significant change in IRF2 protein synthesis (Figure 5B). Further, to reconfirm the actual position of the IRF2 band, western blot was performed with the same IP extracts using antiIRF2 antibody (Figure 5C). The result reconfirms that during IFN treatment, the IRF2 protein level was not significantly reduced. Since, the level of phosphorylated form of eIF2α was found to be increased under similar condition (data not shown), it appears that IRF2-IRES function might continue when cap-dependent translation is largely impaired, suggesting possible role in maintaining intricate balance in the expression of interferon response genes.
To gain further insight on the differential activity of HCV and IRF2-IRES under such condition, we performed UV cross-linking assay using S10 extract isolated from either mock or interferon α-treated Huh7 cells. The results showed a similar protein-binding profile. However, in UV cross-linking experiment p60 binding was found to be much more prominent with the IRF2 5′UTR as compared to HCV IRES and seems to be unaffected with interferon treatment (Figure 5D). It is therefore tempting to speculate that this differential protein binding with the 5′UTRs might influence their respective IRES activity under interferon treatment.
Effect of tunicamycin treatment on the IRF2-IRES activity
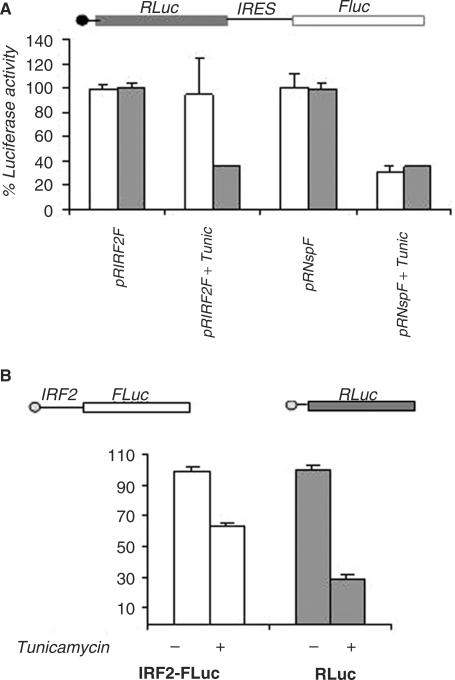
Since, our results suggest that the IRES activity of IRF2 is largely unaffected in presence of interferon α, we wanted to extend our study to investigate the IRES function under ER stress, which is known to cause translational shut down by eIF2α phosphorylation (37). For this purpose, HeLa cells were transfected with either the bicistronic plasmid (pRIRF2F) containing IRF2 5′UTR or the non-specific bicistronic plasmid pRNspF containing a non-specific sequence (as negative control), followed by the treatment with tunicamycin (known to cause ER stress). Interestingly, under such condition, in the context of bicistronic plasmid, the cap-dependent translation of renilla luciferase activity was severely impaired in both pRNspF and pRIRF2F constructs as expected. However, IRF2-IRES-mediated translation of firefly luciferase was not significantly affected (Figure 6A).
Figure 6.
Effect of tunicamycin on the IRES activity of IRF2. (A) HeLa cells were transiently transfected with bicistronic plasmids pRIRF2F or pRnullF. Four hours post-transfection, tunicamycin (Calbiochem) was added at a concentration of 2.5 μg/ml. Luciferase activity was measured 14 h post-transfection. FLuc (white bar) and RLuc (gray bar) activities are shown as fold decrease with respect to corresponding control (without tunicamycin), taken as 100%. The data mean ± SD from three independent experiments. (B) HeLa cells were transiently co-transfected with 1 μg of monocistronic plasmids pCDIRF2FLuc or pCDRLuc. 4 h post-transfection, tunicamycin (Calbiochem) was added at a concentration of 2.5 μg/ml. Luciferase activity was measured 14 h post transfection. FLuc (white bars) and RLuc (gray bars) activities correspond to the activities of the respective reporter constructs (as indicated). FLuc and RLuc activities are shown as fold decrease with respect to corresponding control (without tunicamycin) taken as 100%. The data mean ± SD from three independent experiments.
To further investigate the effect in a closer natural context, we have used monocistronic constructs. In this experiment, monocistronic plasmid pCDIRF2FLuc was used where firefly luciferase reporter gene was cloned downstream of IRF2 5′UTR. As control of cap-dependent translation monocistronic plasmid pCDRLuc was used (Figure 6B). When these plasmids were transfected in HeLa cells, both the monocistronic mRNAs generated were expected to be capped and translated by cap-dependent mode. However, the IRF2-FLuc RNA would have an option to switch to IRES-mediated translation when cap-dependent translation mode is affected due to ER stress (14,38). When pCDRLuc was transiently transfected in the HeLa cells followed by treatment with tunicamycin, the cap-dependent translation of Renilla luciferase activity was found to be inhibited drastically (∼70%) compared to control (Figure 6B). However, only marginal decrease in the firefly luciferase activity (∼30%) was observed in presence of tunicamycin when pCDIRF2FLuc plasmid was used.
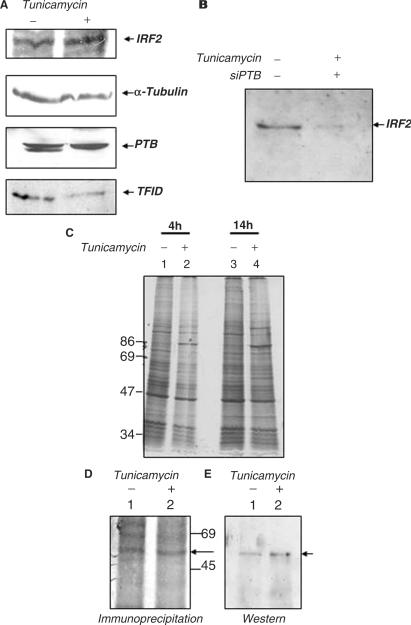
Since we observed that the IRES activity of IRF2 was not significantly altered under tunicamycin treatment, we wanted to study the level of IRF2 protein expression under such condition. Western blot analysis also showed that there was no significant change in the level of IRF2 protein (Figure 7A), although the level of TFIID protein was found to be diminished in presence of tunicamycin (Figure 7A). Interestingly, the level of PTB protein was also not changed under such condition (Figure 7A). We have demonstrated the requirement of PTB protein for IRF2-IRES activity in our earlier assay. Thus to investigate whether availability of PTB could affect the level of IRF2 protein under tunicamycin treatment, we opted for transient silencing of PTB followed by tunicamycin treatment. Equal amount of cell lysates were loaded for western blot analysis using antiIRF2 antibody. The result showed a dramatic decrease in the level of IRF2 under such condition (Figure 7B), suggesting that the availability of PTB is important for the synthesis of IRF2 protein under tunicamycin treatment. The same blot was stripped and probed with anti-beta actin antibody (loading control), which did not show any appreciable difference (data not shown).
Figure 7.
Effect of tunicamycin and transient PTB silencing on IRF2 protein synthesis. (A) 100 μg of untreated and tunicamycin-treated HeLa extracts (as indicated above the panel) were probed with anti- IRF2, anti-α tubulin, anti-PTB and anti-TFIID antibodies. (B) HeLa cells were either mock-treated or treated with 100 nM siPTB. Six hours post siPTB transfection, 2.5 μg/ml of tunicamycin was added to the siPTB-treated cells as indicated above the panel. Extracts were prepared 14 h post tunicamycin treatment and western blot was performed with anti IRF2 antibody. (C) Metabolic labeling of untreated and tunicamycin-treated HeLa cells. HeLa cells were treated with 2.5 μg/ml tunicamycin for the indicated time points and metabolically labeled with 35S-methionine. The extracts were resolved in SDS–8% PAGE gel, followed by autoradiography. Lanes 1 and 3 indicate untreated extracts and lanes 2 and 4 depict tunicamycin-treated extracts. (D) Immunoprecipitation followed by western blot analysis of IRF2 in untreated and tunicamycin-treated extracts. Cells were metabolically labeled in the absence and presence of tunicamycin (as indicated above lanes). Lanes 1 and 2 correspond to immunoprecipitation of IRF2 without and with tunicamycin-treated cell extracts respectively, the arrow indicates the IRF2 protein band.(E) Lanes 1 and 2 correspond to the western blot of IRF2 with the above immunoprecipitated extracts.
Additionally, to monitor IRF2 synthesis we have performed pulse metabolic labeling experiment with the HeLa cells after tunicamycin treatment. This assay would not depend on the stability of the pre-existing protein but will reflect the rate of respective protein synthesis during stress condition. As expected, pulse metabolic labeling of cells treated with tunicamycin for different time periods (4 and 14 h) showed appreciable decrease in overall protein synthesis (Figure 7C). However, immunoprecipitation of IRF2 protein of the above cell extract clearly demonstrated continued synthesis of IRF2 protein (Figure 7D) reconfirming earlier observation. The position of IRF2 band was further confirmed by western blot analysis with the same IP extracts using antiIRF2 antibody (Figure 7E).
Taken together, these observations strongly suggest that IRF2 RNA has an IRES element, which is less sensitive to conditions that lead to shutdown of cap-dependent translation of majority of cellular mRNAs and allow basal level of IRF2 protein synthesis to regulate interferon and other cellular stress response. Also, a trans-acting factor, PTB is important for the IRF2-IRES activity under stress condition and its availability could be an important determinant of the efficiency of the respective IRES activity.
DISCUSSION
Interferon (IFN) stimulates transcription of ‘interferon-stimulated response element (ISRE)’ containing genes by the activation of ‘interferon regulatory factors’ (such as IRF1, IRF7, etc.) (39). This activation also results in increased transcription of these regulatory factors. Thus the effect of global attenuation of translation might not affect their protein level as much. However, it is important to modulate the activity of these IRFs as well to maintain the intricate balance and the cellular response. IRF2 protein has been shown to negatively regulate the interferon pathway. We hypothesize, that continuous synthesis of IRF2 protein by its IRES element probably ensures unaltered protein level under this situation, which in turn might regulate IFN-induced gene expression. In fact western blot analysis did show similar levels of IRF2 protein in control and interferon-treated cells in support of our hypothesis.
In this study, we have shown evidence for a cap-independent translation initiation or IRES-mediated translation of IRF2 that probably allows a response to various stress conditions. Bicistronic assays were employed to show that the IRF2 5′UTR is capable of mediating internal initiation. Furthermore, using stringent assays we have tried to rule out cryptic promoter or splicing activity. Although, our northern blot and RT-PCR analysis clearly showed intact bicistronic RNAs in transfected cells, it is difficult to absolutely rule out the presence of lower abundance of mono-cistronic RNAs generated due to RNA degradation or spurious splicing activity in the context of RLuc/FLuc bicistronic construct as shown in XIAP IRES (40).
In general, IFN causes activation of PKR, which leads to the phosphorylation of eIF2-alpha culminating in the suppression of protein synthesis (2). Our study shows that in the presence of interferon, when majority of protein synthesis is affected, IRF2 is still synthesized by IRES mode of translation. However, this is not true for all cellular IRES, since BiP IRES activity showed significant inhibition. Also HCV IRES was found to be sensitive to interferon treatment as reported earlier (36). Interestingly, analysis of the cellular protein binding with IRF2 and HCV IRES did not show much difference in the profile in absence and presence of IFN-α. However, a 58 kDa polypeptide showed prominent binding with the IRF2 5′UTR only. It is not clear at this stage whether this particular protein is involved in IRF2 IRES function in IFN-treated cells.
Additionally, the IRF2 protein level was also found to be unaltered in tunicamycin-treated cells, when majority of protein synthesis is expected to be inhibited during ER stress due to phosphorylation of eIF2α. This could be explained by the IRF2-IRES activity, which was found to be unaltered during the treatment. The observation put forward the idea that IRF2 protein might be involved in the cellular responses in other stress conditions as well.
Several cellular IRESs have been shown to be active under conditions when eIF4G is cleaved. Incidentally, it has been shown that p50 (a central one-third part of eIF4G) and p100 (C terminal two-third fragment of eIF4G) can partially replace the function of the intact eIF4G in translation initiation mediated by the EMCV IRES (41). In fact, the cleavage product of eIF4G as well as the eIF4G-related protein p97 and its cleavage product p86 has been shown to influence the IRES function (19,42). Interestingly, IRF2 5′UTR showed binding with polypeptides of ∼50 and 100 kDa when 2A protease-treated cell extract was used. Also in our assay, p86 was found to bind with IRF2 but not with HAV IRES. It appears that the binding of these polypeptides could give selective advantage to the IRF2-IRES activity over HAV IRES under conditions of eIF4G cleavage.
PTB protein has been implicated to modulate functions of several cellular mRNAs. It has been shown to act as RNA chaperone and facilitate Apaf1-IRES structure and influence the efficiency of its translation initiation (43). It has also been reported to positively regulate the IRES-mediated translation of HIF-1α, p27kip1, etc (44,45), whereas it negatively regulates unr and BiP IRES function (46,47). Our results suggest that PTB protein might be required for the efficient activity of the IRF2-IRES, since partial knockdown significantly affected the IRES activity. Interestingly, the cytoplasmic pool of PTB protein has been shown to vary under various physiological stress conditions, such as apoptosis and viral infection (48,49). Since it appears that the IRF2-IRES remains active under stress conditions and may be regulatable, it would be interesting to investigate how the abundance of PTB protein during such conditions could influence the IRES function. Currently, experiments are in progress to verify the MFOLD predicted secondary structure of the IRF2-IRES and how PTB binding can influence IRF2-IRES function during physiological stress conditions.
Although, IRF2 protein is known as a negative regulator of interferon-stimulated genes, it has been implicated to stimulate vascular cell adhesion molecule (VCAM) and regulation of histone H4 genes under various conditions (50,51). The fact that the IRF2 gene is translationally regulated under stress conditions (such as ER stress and interferon treatment), it might have indirect effect on other gene expression as well. Thus, it is likely that the regulated expression of IRF2 protein under various stress conditions would have major implications on the cellular response. Incidentally, this study constitutes the first report on translational control of interferon regulatory factors by internal initiation. The results might have far reaching implications on the possible role of IRF2 in controlling the intricate balance of cellular gene expression under stress conditions in general.
ACKNOWLEDGEMENTS
We thank Dr Peter Sarnow (Stanford), Dr Akio Nomoto (Tokyo), Dr Nora Chapman (Nebraska), Dr J.G. Patton, Dr E. Ehrenfeld (NIH) and Dr Bernard Moss (NIH) for plasmid constructs and virus. We gratefully acknowledge the generous gift of antiIRF2-antibody from Dr Angela Battistini of Istituto Superiore di Sanita, Rome, Italy. We also thank Dr C. DurgaRao and Dr K.P. Gopinathan, for providing the eIF4G and TFIID antibodies, respectively. We are also thankful to Dr M.N. Khaja (CLRD, Hyderabad), Dr Raja Murusinge (Genotypic Biotechnology, Bangalore) and Dr P. Ray, Dr M. Venkatramana and other members of our laboratory for their help and suggestions. We thank Dr Umesh Varshney for his critical comments on the manuscript. This work was supported by grant to S.D. from the Department of Biotechnology-Genomics Initiative at IISc. D.D. was supported with pre-doctoral fellowship from Council of Scientific and Industrial Research, India. Funding to pay the Open Access publication charges for this article was provided by S.D. from the Department of Biotechnology-Genomics Initiative at IISc.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nguyen H, Hiscott J, Pitha PM. Growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Oumard A, Hennecke M, Hauser H, Nourbakhsh M. Translation of NRF mRNA is mediated by highly efficient internal ribosome entry site. Mol. Cell. Biol. 2000;20:2755–2759. doi: 10.1128/mcb.20.8.2755-2759.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl Acad. Sci. USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellen CU, Sarnow P. Internal ribosme entry sites in eukaryotic mRNAs. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 6.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrish BC, Rumsby MG. The 5' untranslated region of protein kinase C delta directs translation by an internal ribosome entry segment that is most active in densely growing cells and during apoptosis. Mol. Cell. Biol. 2002;22:6089–6099. doi: 10.1128/MCB.22.17.6089-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoneley M, Chappell SA, Jopling CL, Dickens M, Macfarlane M, Willis AE. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 2000;20:1162–1169. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin X, Sarnow P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- 10.Holcik M, Sonenberg N, Korneluk RG. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 2000;16:469–473. doi: 10.1016/s0168-9525(00)02106-5. [DOI] [PubMed] [Google Scholar]
- 11.Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 12.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol. Cell. Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subkhankulova T, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated initiation of c-myc protein synthesis following genotoxic stress. Biochem. J. 2001;359:183–192. doi: 10.1042/0264-6021:3590183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez J, Yaman I, Sarnow P, Snider MD, Hatzoglou M. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2alpha. J. Biol. Chem. 2002;277:19198–19205. doi: 10.1074/jbc.M201052200. [DOI] [PubMed] [Google Scholar]
- 15.Tinton SA, Schepens B, Bruynooghe Y, Beyaert R, Cornelis S. Regulation of the cell-cycle-dependent internal ribosome entry site of the PITSLRE protein kinase: roles of Unr (upstream of N-ras) protein and phosphorylated translation initiation factor eIF2alpha. Biochem. J. 2005;385:155–163. doi: 10.1042/BJ20040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemens MJ, Bushell M, Jeffrey IW, Pain VM, Morley SJ. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 2000;7:603–615. doi: 10.1038/sj.cdd.4400695. [DOI] [PubMed] [Google Scholar]
- 17.Henis-Korenblit S, Shani G, Sines T, Marash L, Shohat G, Kimchi A. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl Acad. Sci. USA. 2002;99:5400–5405. doi: 10.1073/pnas.082102499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holcik M, Yeh C, Korneluk RG, Chow T. Translational upregulation of X- linked inhibitor of apoptosis (XIAP) increases resistance to radiation induced cell death. Oncogene. 2000;19:4174–4177. doi: 10.1038/sj.onc.1203765. [DOI] [PubMed] [Google Scholar]
- 19.Nevins TA, Harder ZM, Korneluk RG, Holcik M. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 2003;278:3572–3579. doi: 10.1074/jbc.M206781200. [DOI] [PubMed] [Google Scholar]
- 20.Ray PS, Grover R, Das S. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Reps. 2006;7:404–410. doi: 10.1038/sj.embor.7400623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatramana M, Ray PS, Chadda A, Das S. A 25 kDa cleavage product of polypyrimidine tract binding protein (PTB) present in mouse tissues prevents PTB binding to the 5′ untranslated region and inhibits translation of hepatitis A virus RNA. Virus Res. 2003;98:141–149. doi: 10.1016/j.virusres.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Dhar D, Mapa K, Pudi R, Srinivasan P, Bodhinathan K, Das S. Human ribosomal protein L18a interacts with hepatitis C virus internal ribosome entry site. Arch. Virol. 2006;151:509–524. doi: 10.1007/s00705-005-0642-6. [DOI] [PubMed] [Google Scholar]
- 23.Fuerst TR, Niles EG, Studie FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl Acad. Sci. USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- 25.Le SY, Maizel JV., Jr A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Q, Sarnow P. Location of the internal ribosome entry site in the 5' non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Res. 1997;25:2800–2807. doi: 10.1093/nar/25.14.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bert AG, Grepin R, Vadas MA, Goodall GJ. Assessing IRES activity in the HIF-1α and other cellular 5′UTRs. RNA. 2006;12:1–10. doi: 10.1261/rna.2320506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoneley M, Subkhankulova T, LeQuesne JP, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamphear BR, Yan F, Yang D, Waters H-D, Liebig H, Klump E, Kuechler T, Skern T, Rhoads RE. Mapping the cleavage site in protein synthesis initiation factoreIF-4 gamma of the 2A proteases from human Coxsackievirus and rhinovirus. J. Biol. Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- 30.Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 31.Kerekatte V, Keiper BD, Badorff C, Cai A, Knowlton KU, Rhoads RE. Cleavage of Poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 1999;73:709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borman AM, Kean KM. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology. 1997;237:129–136. doi: 10.1006/viro.1997.8761. [DOI] [PubMed] [Google Scholar]
- 33.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 34.Wagner EJ, Garcia-Blanco MA. RNAi mediated PTB depletion leads to enhanced exon definition. Mol. Cell. 2002;10:943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- 35.Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant MJ, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: the next Generation. Gene. 1999;237:1–14. doi: 10.1016/s0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- 36.Kato J, Kato N, Moriyama M, Goto T, Taniguchi H, Shiratori Y, Omata M. Interferons specifically suppress the translation from the internal ribosome entry site of hepatitis C virus through a double-stranded RNA-activated protein kinase-independent pathway. J. Infect. Dis. 2002;186:155–163. doi: 10.1086/341467. [DOI] [PubMed] [Google Scholar]
- 37.Rutkowski TD, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Brewer JW, Diehl JA. PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc. Natl Acad. Sci. USA. 2000;97:12625–12630. doi: 10.1073/pnas.220247197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi T, Takaoka A. A weak signal for strong responses: interferon alpha/beta revisited. Nat. Rev. Mol. Cell. Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 40.Holcik M, Graber T, Lewis SM, Lefebyre CA, Lacasse E, Baird S. Spurious splicing within the XIAP 5′UTR occurs in the Rluc/Fluc but not the betagal/CAT bicistronic reporter system. RNA. 2005;11:1605–1609. doi: 10.1261/rna.2158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 42.Hundsdoerfer P, Thoma C, Hentze MW. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl Acad. Sci. USA. 2005;102:13421–13426. doi: 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosme entry segment attains the correct structural conformation for function with interaction with PTB and UNR. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 44.Schepens B, Tinton SA, Bruynooghe Y, Beyaert R, Cornelis S. The polypyrimidine tract-binding protein stimulates HIF-1alpha IRES-mediated translation during hypoxia. Nucleic Acids Res. 2005;33:6884–6894. doi: 10.1093/nar/gki1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho S, Kim JH, Back SH, Jang SK. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol. Cell. Biol. 2005;25:1283–1297. doi: 10.1128/MCB.25.4.1283-1297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cornelis S, Tinton SA, Schepens B, Bruynooghe Y, Beyaert R. UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein. Nucleic Acids Res. 2005;33:3095–3108. doi: 10.1093/nar/gki611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim YK, Hahm B, Jang SK. Polypyrimidine tract binding protein inhibits translation of bip mRNA. J. Mol. Biol. 2000;304:119–133. doi: 10.1006/jmbi.2000.4179. [DOI] [PubMed] [Google Scholar]
- 48.Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol. Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 49.Back SH, Kim YK, Kim WJ, Cho S, Oh HR, Kim J-E, Jang SK. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract binding proteins executed by polioviral 3C. J. Virol. 2002;76:2529–2542. doi: 10.1128/jvi.76.5.2529-2542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jesse TL, LaChance R, Iademarco MF, Dean DC. Interferon regulatory factor 2 is a transcriptional activator in muscles where it regulates expression of vascular cell adhesion molecule-1. J. Cell Biol. 1998;140:1265–1276. doi: 10.1083/jcb.140.5.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaughan PS, van der Meijden CM, Aziz F, Harada H, Taniguchi T, van Wijnen AJ, Stein JL, Stein GS. Cell cycle regulation of histone H4 gene transcription requires the oncogenic factor IRF2. J. Biol. Chem. 1998;273:194–199. doi: 10.1074/jbc.273.1.194. [DOI] [PubMed] [Google Scholar]