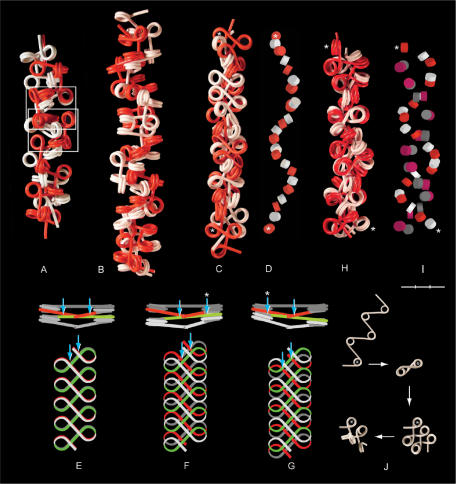
Figure 5.
Four types of chromatin double fibers formed by polygon fibers (red and white). (A) Two filaments with β sequence (333)n and α = 108° (as shown in Figure 4B) align and interlock like a zip, in which nucleosomes in one fiber fit spatially into the space provided by a swivel-linker between two trinucleosomes in the other fiber (framed). (B) Two fibers with closed, negative linker configuration, β sequence (3033)n, α = 108°, coil right-handed around each other forming a double helix. The swivel-linkers are parallel to the axis of the fiber, with alternating positions of the red and the white filament on the outside of the fiber. (C) Two fibers coil left-handed around each other by intercalation of every second nucleosome, α varying between 85 and 120° and β between 72 and 160°. (D) The positions of the intercalated core particles in the fiber in (C). (E–G) Two nucleosomal filaments with β(5) (180°) placed one on top of the other. (E) Nucleosomes stacking face-to-face with every second linker (red and green) touching each other at two crossover points (arrows), which are symmetrically positioned relative to the midpoint of the linker. (F and G) Parallel staggering of the ribbons along the fiber axis causes one crossover to be displaced toward the middle of the linker and the other toward the nucleosomal DNA of every second nucleosome, which is located at one edge of the ribbon (arrows). (H and I) A left-handed double helix formed by two filaments with alternating negative and positive linker configuration folded from a filament with open linker configuration; ∼90° < α < ∼108°; ∼β(+3). (I) The positions of the core particles in (H) with negative (red and white) and positive linker configuration (mauve and gray). (J) Folding of a nucleosomal filament leading to the conformation in (H). The first nucleosome is marked by an asterisk. Asterisks in (C, D, H, I) indicate the first and the last nucleosome in a sequence of intercalated nucleosomes. Bar = 30 nm (A–I).