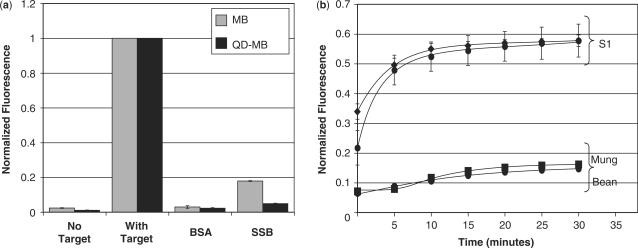
Figure 6.
Fluorometric analysis of MB- and QD–MB-protein interactions and endonuclease degradation. (a) MBs and QD–MBs were incubated with molar excess of complementary target, BSA and SSB (n = 3 each) and the peak fluorescent intensity of the MB was measured. SSB generated a false-positive signal upon binding to MBs and QD–MBs, although the increase in signal was much less pronounced with QD–MBs. (b) Incubation of MBs and QD–MBs with S1 endonucleases resulted in a significant increase in fluorescence. Both probes were degraded at similar rates. Mung Bean nuclease was also capable of degrading MBs and QD–MBs but to a much lesser extent than S1 nucleases. All fluorescent measurements were normalized to the signal elicited by MBs and QD–MBs, respectively, in the presence of saturating concentrations of complementary target.