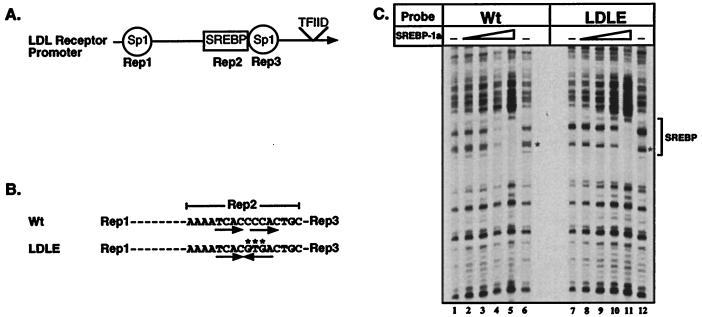
Figure 1.
Wild-type and mutant LDL receptor promoters: Structure and DNase I footprint analysis by SREBP-1a. A schematic view of the region of the LDL receptor promoter required for sterol regulation containing repeats 1–3 (A) and a more detailed view of the sequence of the wild-type (LDL) and mutant repeat 2 (LDLE) are shown (B). The mutated bases are indicated by ∗ and the direct repeat and palindromic motifs are indicated by the arrows. (C). DNA probes end-labeled with 32P on the top strand were prepared from plasmids containing the wild-type (Wt, lanes 1–6) or mutant (LDLE, lanes 7–12) promoters. DNA probe (0.01 pmol) were used in each binding reaction in the absence or presence of increasing amounts (0.02, 0.06, 0.2, and 0.6 pmol) of recombinant SREBP-1a (amino acids 1–490). DNase I footprinting was performed as described in Materials and Methods. The asterisk on the autoradiogram (C) indicates the position of differences in the observed cleavage pattern of free DNA due to the 3-bp difference between the two probes. The minimal size of the DNase I footprint is indicated by the bracket at the right of the figure.