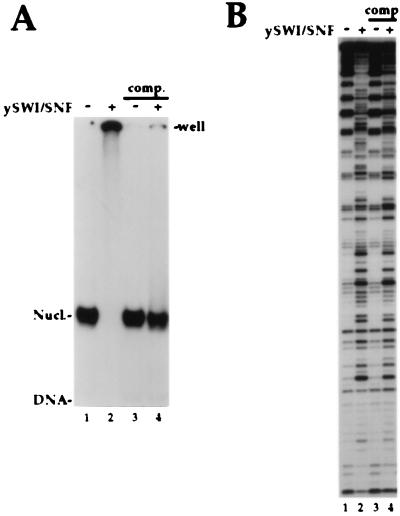
Figure 2.
Dissociation of the SWI/SNF complex leaves a persistently altered nucleosome core. (A) A 5S DNA probe as in Fig. 1 was reconstituted into nucleosome cores. SWI/SNF binding was analyzed by standard gel shift assay [4% acrylamide (29/1)/0.5× TBE]. Each reaction contains 12.5 nM total nucleosomes and 1 mM Mg-ATP, whereas 30 nM SWI/SNF was added in lanes 2 and 4. In lanes 3 and 4, after an initial 30-min incubation, 20-fold excess of cold HeLa oligonucleosomes were added followed by another 30-min incubation at 30°C before loading on the gel. (B) Binding reactions similar to A were performed and analyzed by DNase I footprint assay. 15 nM of SWI/SNF was added in lanes 2 and 4. Competitors and 2-fold more DNase I were used in lanes 3 and 4. Near stochiometric ratio of SWI to nucleosomes were used here and in the following experiments to insure almost complete binding of the nucleosomes in the initial reaction.