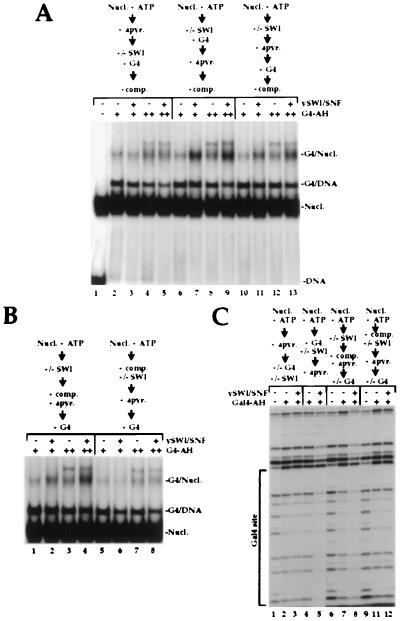
Figure 3.
Temporal analysis of SWI/SNF action and enhanced activator binding. (A) SWI/SNF-treated nucleosome cores remain bound by GAL4-AH after SWI/SNF detachment. A 154-bp DNA probe bearing a single GAL4 site 32 bp from one end was reconstituted into nucleosome cores and incubated with 30 nM (lanes 2, 3, 6, 7, 10, and 11) and 100 nM (lanes 4, 5, 8, 9, 12, and 13) GAL4-AH dimers. All lanes contain 12.5 nM total nucleosomes and 1 mM ATP, and 10 nM SWI/SNF was included in the odd-numbered lanes. To study the temporal ATP requirement for SWI action, ATP was degraded by apyrase before SWI/SNF addition (lanes 2–5), after SWI/SNF and GAL4 incubation (lanes 6–9), or after SWI/SNF action but before GAL4 incubation (lanes 10–13). Finally, before loading the native gel, nucleosome cores were released from SWI by competition with 20-fold excess of cold oligonucleosomes for 30 min at 30°C. (B) SWI/SNF treatment of nucleosome cores allows GAL4-binding after SWI/SNF detachment. Gel shift assay of similar binding reactions as in A except for the time of SWI/SNF competition. In lanes 1–4 competitors were added after SWI/SNF action, at the same time as apyrase treatment, before GAL4 addition. In lanes 5–8 competitors were included from the beginning of the binding reaction, at the same time as SWI/SNF is added. (C) Footprinting assay of equivalent binding reactions. 30 nM GAL4-AH dimers was added in lanes 2–5 whereas 600 nM was used in lanes 7, 8, 11, and 12. SWI/SNF (40 nM) was added at the indicated time in lanes 3, 5, 8, and 12. Competitors were added at the indicated time in lanes 6–12. Two-fold more DNase I was used in lanes 6–12.