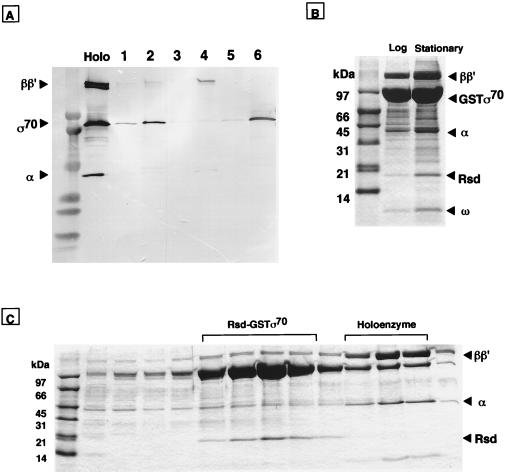
Figure 3.
Identification of complex formation between σ70 and Rsd. (A) Complex formation in vitro between Rsd and σ70 was analyzed by the GST pull down assay. GST (lanes 1, 3, and 5) or GST–Rsd (lanes 2, 4, and 6) was mixed with equimolar amounts of Eσ70 holoenzyme (lanes 1 and 2), core enzyme (lanes 3 and 4), or σ70 subunit (lanes 5 and 6). Complexes formed were bound to glutathione–Sepharose beads. The bead-bound proteins were eluted with 50 mM glutathione and separated by SDS/PAGE on a 5–15% gradient gel. The gel was subjected to Western blot analysis by using a mixture of monospecific antibodies against RNA polymerase α, β, β′, and σ70 subunits. The migration positions of RNA polymerase subunits can be identified in the holoenzyme lane. (B) Isolation of GST–σ70-associated proteins in cell extracts. Cell lysates of a pGEXD transformant of E. coli W3110 were prepared at both exponential (lane, log) and stationary (lane, stationary) phases. GST–σ70-bound proteins were isolated by the GST pull down assay, and the σ70-bound proteins were separated by SDS/13.5% PAGE. The migration positions of core enzyme subunits, GST–σ70, and ω proteins are indicated on the right. (C) Proteins isolated from stationary phase cells (see stationary lane in B) were subjected to heparin–Sepharose column chromatography. Proteins were eluted by a linear gradient of NaCl, and aliquots were analyzed by SDS/13.5% PAGE.