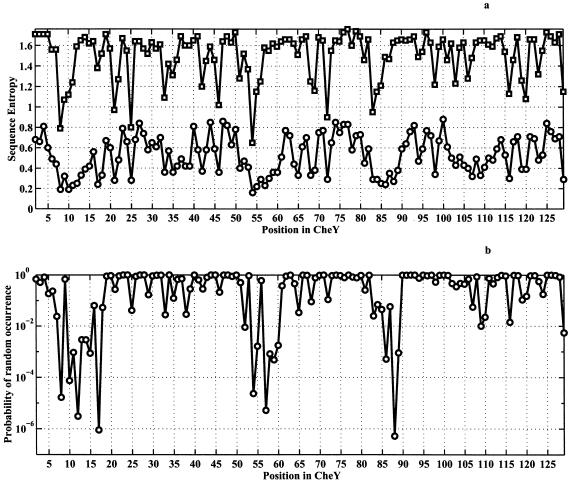
Figure 4.
Analysis for the CMBF superfamily. Ninety-eight families were used (the list is available from authors on request). All listed proteins are structurally homologous to CheY with Z > 3 and RMSD < 4A, according to the families of structurally similar proteins (FSSP) database (19). We used a coarse-grained six-letter amino-acid alphabet whereby amino acids were grouped according to their physical properties into following six classes: “aliphatic + Cys”: A, L, I, V, M, C; “aromatic”: F, Y, W, H; small nonpolar: G, P; polar: T, S, Q, N; basic: R, K; and acidic: E, D. The analysis using all 20 types of amino acids gives results that are qualitatively similar. Horizontal axes denote position in the CheY, which was taken as reference. (a, circles) CoC analysis: intrafamily sequence entropy averaged over all 98 families (excluding gaps), calculated as SCoC(l) = ∑F=1M SintraF(l)/M. Here, the sum is taken over all of the 98 families used in the analysis, excluding gaps. Intrafamily sequence entropy for every position, for a given family, F, is calculated as follows: SintraF(l) = −∑i=16 piF(l)log piF(l), where piF(l) represents the normalized frequency of observing residue of class i (i = 1–6) at position l in all homologous sequences belonging to the family F. The sum is taken over all possible residue classes. (a, squares) sequence entropy calculated across all families. To obtain this quantity, we evaluated frequencies of occurrence of amino acids of each class i at each position l for all families [piacross(l)] and then calculated sequence entropy for a position l as Sacross(l) = −∑i=16 piacross(l)log piacross(l). (b) The probability that equal or lower SCoC will be observed under zero hypothesis that conservatism of a residue in the structure is related primarily to its degree of buriedness.