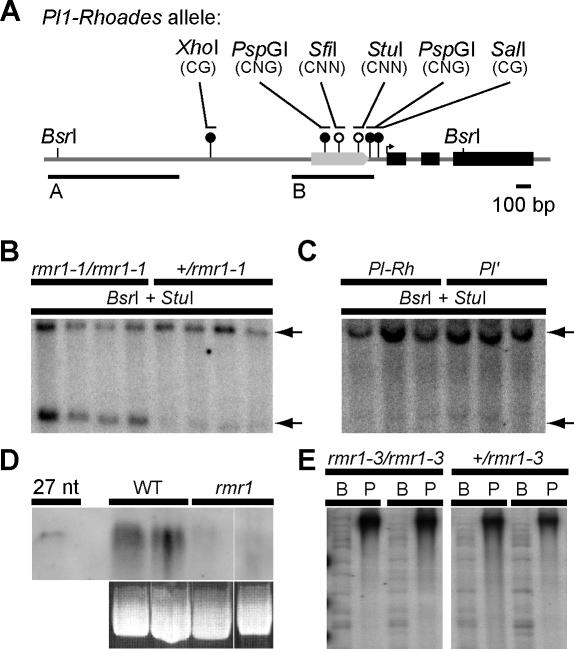
Figure 4. Cytosine Methylation Patterns and Small RNA Accumulation Are Altered at Pl1-Rhoades in rmr1 Mutants.
(A) Schematic of Pl1-Rhoades locus with exons highlighted in black and the upstream doppia element represented by the gray arrow. The methylation context of sites cut by methylation-sensitive enzymes are shown in parentheses. Open circles denote sites hypomethylated in rmr1–1 mutants while filled circles are sites methylated in both wild-type and rmr1 mutants. BsrI restriction sites and the regions used to generate probes for blot hybridization analysis, denoted A and B, are also shown.
(B and C) Representative Southern blots hybridized with probe A showing methylation status at a StuI site in rmr1 mutants and heterozygous siblings (B), as well as Pl′ and Pl-Rh plants (C) with a larger 2.9-kb band (upper arrow) representative of a fully methylated BsrI fragment, and a 2.1-kb band (lower arrow) indicative of a hypomethylated StuI site. Additional primary blots shown in Figures S3 and S8.
(D) Small RNA northern blot probed with doppia sequence from probe B showing changes in amount of small RNAs between rmr1–1 plants and wild-type (WT) siblings.
(E) Southern blot of genomic DNA digested with BstNI (“B” lanes) and methylation-sensitive PspGI (“P” lanes) hybridized with probe B, showing no bulk changes in doppia methylation genome-wide.