Figure 1.
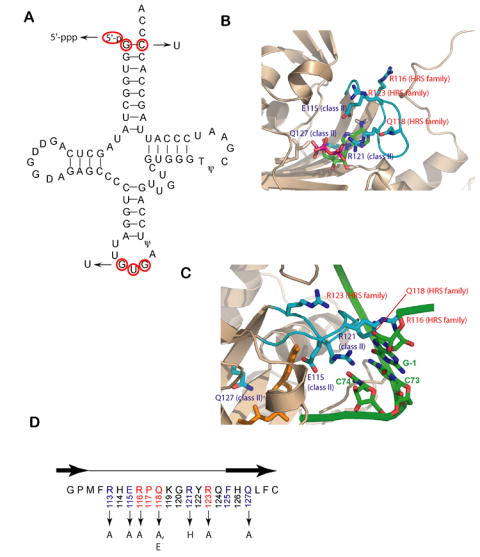
tRNA recognition elements in the HisRS/tRNAHis system.
(A) Cloverleaf diagram of tRNAHis from E. coli. Key recognition elements for tRNAHis by HisRS are shown in red. The mutant substitutions used in this work are denoted by arrows.
(B) Close up of the active site of the E. coli HisRS histidinol-ATP complex, showing residues of the motif 2 loop in contact (PDB id: 1KMN). The motif 2 loop and residues thereof are colored in cyan, while the surrounding active site secondary structures are rendered in tan. Class II conserved residues are labeled in blue, while the HisRS-family conserved residues are labeled in red.
(C) Interactions between HisRS and tRNAHis predicted by homology modeling with the AspRS-tRNAAsp crystal structure (Connolly et al., 2004). The coloring of the enzyme is as noted in panel A, while the nucleotides of tRNAHis are colored green. The aminoacyl-adenylate is shown in orange. The model has not been manipulated to reflect conformational rearrangements likely associated with tRNA binding. The configuration of the motif 2 loop represents the “T state” seen in one of the two monomers of the histidyl adenylate complex (PDB: 1KMM).
(D) Primary sequence of the E.coli HisRS motif 2 loop. The β strands flanking loop are depicted above the sequence as black arrows. Class II conserved amino acids are highlighted in light blue, while HisRS conserved sequences are highlighted in red. Amino acid substitutions in the motif 2 loop examined in this study are indicated by arrows.
