Fig. 1.
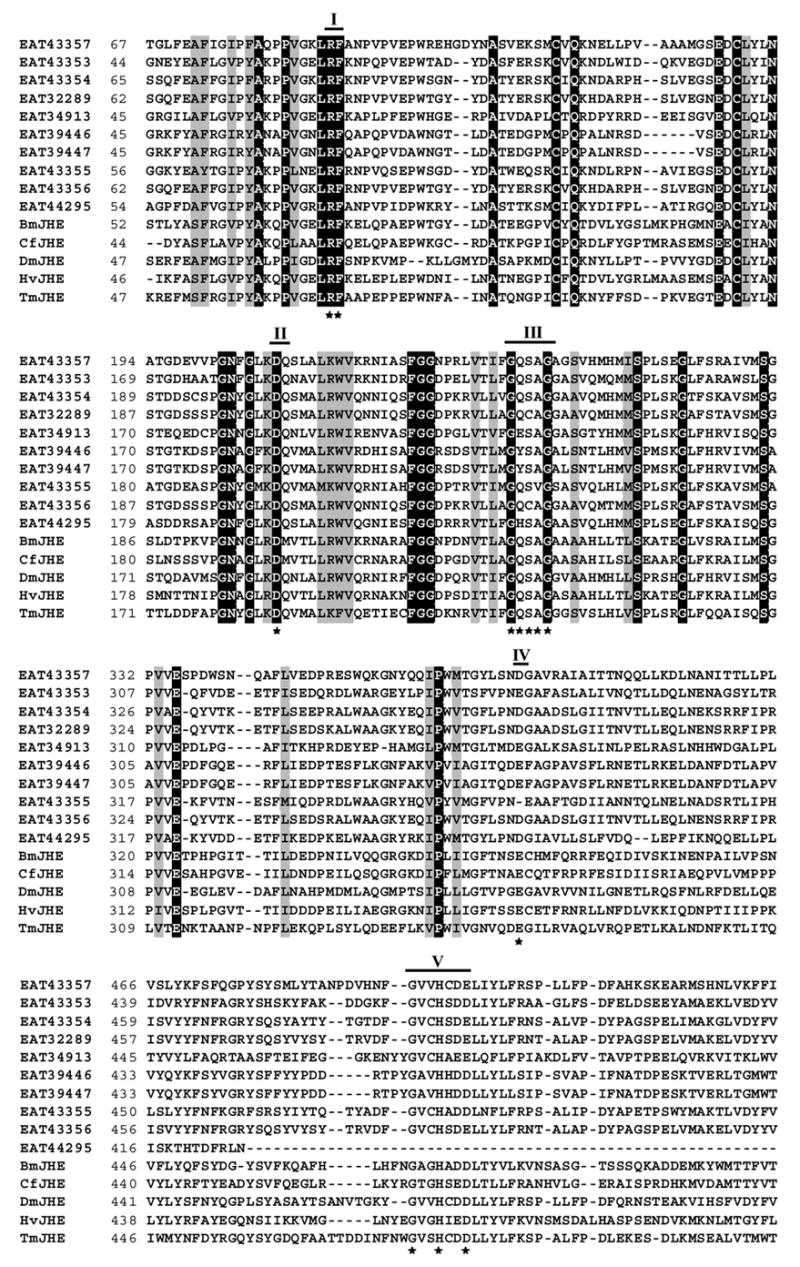
Alignment of amino acid sequences of ten putative jhe genes from Aedes aegypti, and jhe genes of Drosophila melanogaster (DmJHE, Accession # NM_079034), Choristoneura fumiferana (CfJHE, Accession # AAD34172), Tenebrio molitor (TmJHE, Accession # AAL41023), Heliothis virescens (HvJHE, Accession # AAC38822), Bombyx mori (BmJHE, Accession # AAR37335), using ClustalW program (http://www.ebi.ac.uk/clustalw) and BioEdit Sequence Alignment Editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Five conserved catalytic motifs are indicated by bold lines with the numbers on the top. Numbers on the left show the position of amino acids in each protein. Letters with black and gray shading denote perfect and similar identities, respectively. The motifs important for JHE function are shown as stars. GENSCAN program (http://genome.dkfz-heidelberg.de/cig-bin/GENSCAN/genscan.cgi) was used to predict the full length ORF (547 aa) of EAT43353 gene
