Abstract
Apurinic/apyrimidinic (AP) endonuclease (APE; EC 4.2.99.18) plays a central role in repair of DNA damage due to reactive oxygen species (ROS) because its DNA 3′-phosphoesterase activity removes 3′ blocking groups in DNA that are generated by DNA glycosylase/AP-lyases during removal of oxidized bases and by direct ROS reaction with DNA. The major human APE (APE-1) gene is activated selectively by sublethal levels of a variety of ROS and ROS generators, including ionizing radiation, but not by other genotoxicants—e.g., UV light and alkylating agents. Increased expression of APE mRNA and protein was observed both in the HeLa S3 tumor line and in WI 38 primary fibroblasts, and it was accompanied by translocation of the endonuclease to the nucleus. ROS-treated cells showed a significant increase in resistance to the cytotoxicity of such ROS generators as H2O2 and bleomycin, but not to UV light. This “adaptive response” appears to result from enhanced repair of cytotoxic DNA lesions due to an increased activity of APE-1, which may be limiting in the base excision repair process for ROS-induced toxic lesions.
Reactive oxygen species (ROS), including partially reduced oxygen species, namely, superoxide anion (O2·–), H2O2, and the hydroxyl radical (⋅OH), are generated endogenously as by-products of respiration and also during pathological states in activated neutrophils (1–3). ROS can also be generated by external agents, such as ionizing radiation (4) and long-wavelength UV light (5) and during binding of transition metals and other ligands such as tumor necrosis factor α and interleukin 1 (6). Because of their ubiquitous and continuous presence, these reactive radicals and oxidizing agents have been implicated in the etiology of a variety of diseases, including arthritis, cancer, and neurodegenerative diseases, and also in aging (7–9). While ROS react with most cellular components, they are also invariably genotoxic, because they react with both the deoxyribose and bases in DNA, and so generate a plethora of base lesions and strand breaks (4, 10, 11). Many of these lesions are cytotoxic and mutagenic (12, 13). Most, if not all, of the lesions induced by ROS are repaired via the base excision repair (BER) pathway, in which the damaged bases are removed by specific DNA glycosylases, leaving abasic sites that are subsequently cleaved by apurinic/apyrimidinic (AP) endonucleases [APEs; EC 4.2.99.18 (14, 15)]. The major mammalian APE (called APE-1) contributes more than 80% of the total APE activity and is analogous to Escherichia coli exonuclease III (Xth protein) (16–19). The majority, if not all, of the ROS-altered base lesions in DNA are removed by two DNA glycosylases–endonuclease III (NTH) and 8-oxoguanine-DNA glycosylase (OGG)—in both E. coli and mammalian cells (20–25). These enzymes have a second activity, AP-lyase, which in a concerted reaction cleaves the base N-glycosyl bond, and carries out β or βδ elimination of the resulting free deoxyribose. They thus cleave the phosphodiester bond 3′ to the abasic site and generate 5′-phosphate and 3′-phospho-α,β-unsaturated aldehyde or 3′-phosphate termini (14). The 3′ blocked ends are not primers for DNA polymerases, so the blocking groups have to be removed before repair synthesis is initiated. APE, in addition to AP site-specific endonuclease activity, possesses a DNA 3′-phosphoesterase activity and generates 3′-OH termini by removing these 3′ blocking groups as well as DNA 3′-phosphoglycolaldehyde produced by ROS-induced DNA strand cleavage (4). Thus, because of its dual activity, APE plays a central role in BER by providing a 3′-OH primer for repair synthesis of DNA after all types of oxidative damage (26).
The relative endonuclease vs. phosphoesterase activity varies widely in APEs from different sources. E. coli Xth has a relatively strong phosphoesterase activity (17). However, the mammalian APEs have very low 3′-end-removing activity (17, 19, 27). Our recent studies on repair of ROS-induced DNA strand breaks by human cell-free extracts indicate that the major APE was primarily responsible for this 3′-end-cleaning activity and was rate-limiting in the repair process (28).
In view of the critical role of APE in repair of oxidative DNA damage, it is important to ask whether APE is regulated by ROS in mammalian cells. Bacteria, notably E. coli, exhibit an “adaptive response” in which sublethal exposure to genotoxic agents, including alkylating agents and ROS, induces specific regulons that encode sets of enzymes involved in inactivating either the agent itself or its reactive species, in ameliorating their effects, or in repair of DNA damage induced by such agents (28–31). Specifically, ROS induce a variety of genes in E. coli that are responsible for countering the oxidative damage. These include endonuclease IV, an APE, which along with Xth removes 3′ blocking groups that are induced in DNA by ROS (4). Although there are reports of adaptive responses in mammalian cells (32–34), no DNA repair enzyme has been directly implicated in these responses. Therefore, we examined whether mammalian cells show a similar adaptive response to ROS. We now report that APE-1 is activated selectively by nontoxic levels of a variety of ROS, and that this transient activation is correlated with increased cellular resistance to oxidizing agents.
MATERIALS AND METHODS
Cell Culture and Cell Survival Assay.
Human HeLa (HeLa S3; ATCC CCL 2.2) cells and WI 38 primary lung fibroblasts, and the V79 Chinese hamster lung fibroblast line from the American Type Culture Collection were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM)/F-12 (1:1) (GIBCO/BRL), supplemented with 10% fetal bovine calf serum (FBS; GIBCO/BRL). All cultures were routinely tested for mycoplasma contamination by using Hoechst stain 33258. For clonogenic survival assays, exponentially growing HeLa or V79 cells were seeded at a density of 200 cells per 60-mm Petri dish in the presence of 0.5% FBS, and 3 h later were treated with 130 nM HOCl. After 12 h of incubation cells were treated with increasing doses of genotoxic agents [H2O2, methyl methanesulfonate (MMS), bleomycin, and HOCl] for 1 h, by adding stock solutions directly to the medium, or cells were exposed to x-rays or UV light. The cells were washed with PBS and then grown in medium containing 10% FBS until the colonies reached ≈1 mm in diameter (10–12 days). The fraction of surviving cells relative to the original number of cells was calculated and then normalized to the zero-dose plating efficiency to determine the surviving fraction at each dose.
Assay of APE Activity in Cell-Free Extracts.
Whole cell extracts were prepared from HeLa cells treated with 0 or 130 nM HOCl (Sigma) according to a published method (35). Aliquots (10 ng) of the extracts were incubated with a 5′-32P-labeled 43-mer duplex oligonucleotide (250 fmol) containing a tetrahydrofuran residue at position 31 (36), in a 50-μl reaction mixture (66 mM Tris⋅HCl, pH 8.0/1 mM MgCl2/0.2 mM EDTA/1 mM dithiothreitol, and 100 μg/ml BSA). After incubation for various times, aliquots were analyzed by electrophoresis in 20% polyacrylamide gels containing 8 M urea in Tris/borate/EDTA buffer. The radioactivity in the 30-mer cleaved products was quantified with a model 425 PhosphorImager (Molecular Dynamics).
Analysis of Gene Expression at the RNA and Protein Levels.
For Northern blot analysis, a 15-μg aliquot from each RNA sample extracted from cells was fractionated, transferred to a nylon membrane, and then hybridized (37) with 32P-labeled cDNAs (19) for APE-1 mRNA or 18S ribosomal RNA. Hybridization signals were recorded by exposure on x-ray films or quantified by using imagequant (Molecular Dynamics) software.
Nuclear run-on assays were carried out as previously described (37, 38). Purified nuclei obtained from mock- and HOCl-treated logarithmic-phase HeLa cells were incubated in [α-32P]UTP-containing transcription assay mixture and then the total RNA was extracted (38). RNA aliquots containing equal amounts of radioactivity were hybridized (37) with immobilized denatured APE-1 cDNA or β-actin cDNA (19). The relative abundance of APE-1 mRNA in each sample normalized to β-actin mRNA was determined from the autoradiogram of the blot.
For Western blot analysis, equal amounts of total cellular protein from different samples were separated by SDS/PAGE (10% polyacrylamide), blotted onto nitrocellulose membrane (Trans-Blot 0.2 μm; Bio-Rad), and then exposed to affinity-purified rabbit antiserum raised against recombinant human APE-1 (hAPE-1). The APE-1 bands were visualized according to the ECL (enhanced chemiluminescence) Western blotting protocol (Amersham).
For intracellular localization of APE-1, HeLa and WI 38 cells, cultured on microscope coverslips, were fixed and then exposed to 1:100 affinity-purified anti-hAPE-1 rabbit antibody. The binding of primary antibody was visualized by using fluorescein-conjugated anti-rabbit secondary antibody (Organon Teknika Cappel) in a Zeiss UV microscope.
Irradiation of Cells.
Cells in 60-mm dishes were irradiated with γ-rays from a 137Cs irradiator (JE Shepard and Associates) with a dose rate of 6.7 Gy/min at room temperature and immediately transferred to the incubator. For UV irradiation, the culture medium was removed from the Petri dish and the cells were irradiated with UVC (254 nm) at 0.5 J/m2. After irradiation, the medium was added back to the cells for further incubation.
RESULTS
Induction of the APE-1 Gene by Sublethal Doses of ROS.
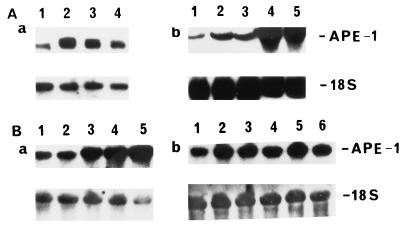
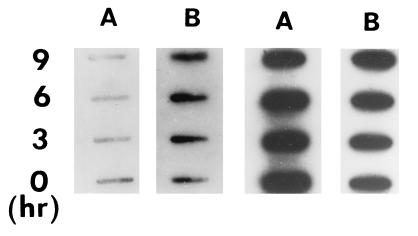
HOCl, a ROS synthesized from H2O2 and Cl− by myeloperoxidase released from activated neutrophils, is a strong oxidant (1, 3). Low doses (130, 260, 850 nM) of HOCl, which showed no detectable cellular toxicity, caused a 5- to 10-fold increase in the level of APE-1 mRNA in logarithmic-phase HeLa cells (Fig. 1Aa). When 130 nM HOCl was used, the maximal APE-1 level was observed 9–12 h after treatment (Fig. 1Ab). This increase in APE-1 mRNA was due to de novo synthesis, because actinomycin D (10 μg/ml), but not anisomycin (a protein synthesis inhibitor) abrogated the increase in APE-1 RNA (Fig. 3A, lanes 3 and 4). Further support for this conclusion was provided by nuclear run-on experiments in which nuclei from mock- or 130 nM HOCl-treated HeLa cells were isolated at various times after treatment and incubated with radiolabeled NTPs for elongation of nascent RNA chains (38). Total RNAs from these samples were then extracted and used as probes in slot-blot hybridization to quantitate APE-1-specific mRNA. It is evident from Fig. 2 Left that the level of nascent APE-1 mRNA increased about 6-fold in HOCl-treated cells (lane B) but not in control cells (lane A). No significant change in the level of β-actin RNA, used as the internal control, was observed in either control or HOCl-treated cells (Fig. 2 Right, lanes A and B).
Figure 1.
(A) Activation of the APE-1 gene in HeLa cells by HOCl. (a) Logarithmic-phase HeLa cells were treated with 0 (lane 1), 130 (lane 2), 260 (lane 3), or 850 (lane 4) nM HOCl for 1 h, and then fed with fresh medium. After 9 h, total RNA was extracted and Northern blot analysis was performed with APE-1 cDNA probes. (b) Kinetics of APE-1 mRNA activation. HeLa cells were treated with 130 nM HOCl, and at 0, 3, 6, 9, and 12 h after treatment (lanes 1–5, respectively), the cells were harvested from individual dishes for RNA analysis as in a. (B) Activation of the APE-1 gene by γ-rays. (a) Effect of radiation dose. Logarithmic-phase HeLa S3 cells were exposed to 0, 1, 2, 4, or 8 Gy of radiation 9 h before cells were harvested and RNA was extracted for Northern blot analysis (lanes 1 through 5, respectively). (b) Kinetics of APE-1 mRNA activation. HeLa cells were exposed to 2 Gy of γ-rays and harvested 0, 3, 6, 9, 12, or 18 h after irradiation. RNA from these cells was loaded for Northern analysis in lanes 1 through 6, respectively. All blots were reprobed for 18S ribosomal RNA (Lower), which was used as an internal control to correct for unequal RNA loading.
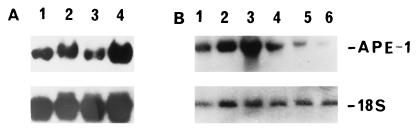
Figure 3.
Effect of inhibitors on HOCl-mediated APE-1 mRNA induction. (A) After treatment with 130 nM HOCl for 9 h, aliquots of HeLa cells were exposed to no drug (lane 2), 5 μg/ml actinomycin D (lane 3), or 100 μg/ml anisomycin (lane 4) for 180 min before extraction of RNA, which was then used for the Northern blot analysis. Lane 1, APE-1 mRNA from control (without HOCl treatment) cells. (B) Abrogation of APE-1 activation by protein kinase inhibitor or ROS scavenger. Logarithmic-phase HeLa cells were mock-treated (lane 1) or treated with 65 nM HOCl (lane 2), 130 nM HOCl (lane 3), simultaneously with 130 nM HOCl and 20 mM N-acetylcysteine (lane 4), or 10 nM staurosporine (lane 5), or 20 nM H7 (lane 6). Northern blot analysis was performed after 12 h of treatment. The blots were reprobed for 18S ribosomal RNA (Lower) as internal control as in Fig. 1.
Figure 2.
Nuclear run-on analysis of APE-1 transcription. Untreated cells (lanes A) and cells treated with 130 nM HOCl (lanes B) for various times were harvested and their nuclei were isolated by a mild detergent lysis procedure (37, 38). Aliquots of nuclei were incubated for 30 min at 30°C with a transcription mixture containing [α-32P]UTP, and the RNA was extracted by using RNAzol (Tel-Test, Friendswood, TX). The extracted RNA was hybridized to a nitrocellulose membrane on which 1 μg of APE-1 cDNA (Left) or β-actin cDNA (Right) was loaded in individual slots.
The specificity of APE-1 induction by ROS was further assessed by using a ROS scavenger, N-acetylcysteine (39) and inhibitors (H7, staurosporine) of protein serine/threonine kinases that were previously shown to be involved in ROS-mediated signaling (6). Fig. 3B shows that treatment of HeLa S3 cells with N-acetylcysteine (20 mM; Fig. 3B, lane 4) or H7 (20 nM; Fig. 3B, lane 6) or staurosporine (10 nM; Fig. 3B, lane 5) decreased APE-1 mRNA.
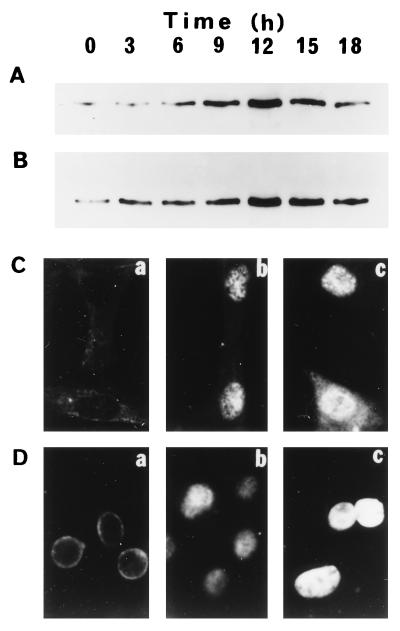
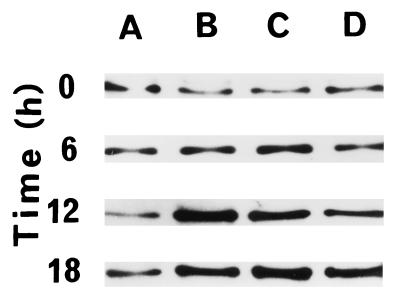
Western blot analysis was undertaken to investigate changes in APE-1 at the protein level. We have consistently found a 3- to 8-fold increase in APE-1 level in HeLa cells (Fig. 4B) and similar increases in additional cell lines (e.g., HepG2, U937, V79; data not shown) and in WI 38 cells (Fig. 4A) after treatment with HOCl (130 nM).
Figure 4.
Activation and intracellular localization of APE-1 in HeLa and WI 38 cells. Logarithmic-phase cells were treated with 130 nM HOCl for 1 h, and at indicated times cells were fixed for immunofluorescence assay or harvested for Western blot analysis. (A and B) Western blot analysis of APE-1 activation in WI 38 (A) and HeLa (B) cells at various times and analyzed for APE-1 level. (C and D) Intracellular localization of APE-1 in WI 38 (C) and HeLa (D) cells, at 0 h (a), 6 h (b), and 12 h (c) after treatment with 130 nM HOCl. APE-1 was visualized by indirect immunofluorescence.
No cellular toxicity was observed even at the highest concentration (850 nM) of HOCl, as described later (Fig. 6). HOCl toxicity was also monitored by flow cytometry, in which no difference was observed in the cell cycle distribution between control and 260 nM HOCl-treated cells up to 24 h after oxidant treatment (data not shown). We found that there was no detectable cell cycle arrest or delay in cell cycle progression for HOCl-treated cells relative to nontreated ones (data not shown). Furthermore, Western blotting analysis indicated that the levels of cell cycle-regulating proteins such as p21 and p53 were not enhanced by HOCl treatment (data not shown).
Figure 6.
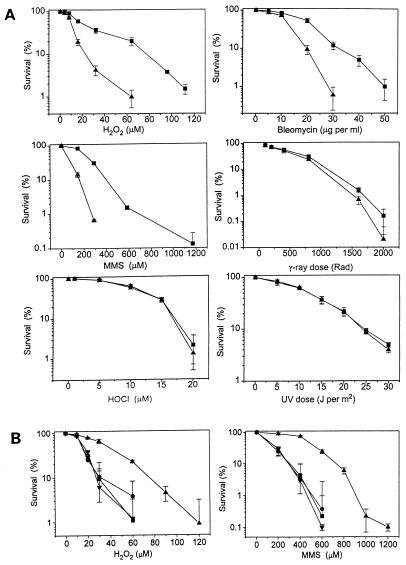
Adaptive response of HeLa cells to oxidative stress. Logarithmic-phase cells (200 per dish) were seeded on 60-mm culture dishes. Three hours later, cells were treated with 130 nM HOCl for 1 h. (A) After 12 h, the cells were exposed to various concentrations of H2O2, bleomycin, MMS, or HOCl, or to various doses of γ-rays or and 254-nm UV light. The untreated (control) and treated cells were fed with fresh medium and allowed to grow for 12–14 more days and the cell colonies (>1 mm) were counted. Triplicate sets of dishes were used for each point and the experiment was repeated three to five times. Colonies were counted and percent of survival was corrected by cloning efficiency and expressed as the mean ± SE. ▪, HOCl-treated; ▴, mock-treated. (B) HOCl- and mock-treated cells were exposed to H2O2 or MMS at 12 h or 24 h after treatment. Cell survival was assayed as in A. Results are for H2O2 or MMS at 12 h (▴) or 24 h (•) after HOCl treatment and mock-treated cells exposed to H2O2 or MMS at 12 h (▪) or 24 h (▾).
ROS Mediates APE-1 Accumulation in the Nucleus.
As a DNA repair protein, the APE-1 polypeptide is expected to be localized in the cell nucleus, as was found to be the case in earlier studies (40, 41). Furthermore, APE-1 has two other unrelated activities, as a reductive activator of AP-1 transcription factors and as a repressor of the human parathyroid hormone gene, that also warrant its nuclear presence (42, 43). Western blot analysis showed a time-dependent increase in APE-1 levels in both WI 38 (Fig. 4A) and HeLa (Fig. 4B) cells. These values reached a maximum at 12 h after treatment with 130 nM HOCl, then declined. Immunofluorescence studies with APE antibody showed that the endonuclease was distributed in the nucleus and cytoplasm of WI38 cells (Fig. 4Ca) and in the cytoplasm and nuclear membrane of HeLa cells (Fig. 4Da). Concomitant with increase in the absolute level at 12 h after HOCl, most of the APE-1 was found to be translocated to the nucleus in both cell types (Fig. 4 Cc and Dc). Thus specific translocation to the nucleus appears to be mediated by ROS signaling.
Activation of APE Mediates Enhanced DNA Damage Repair.
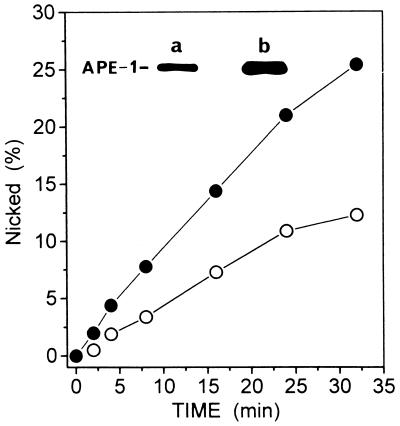
In view of the recent studies indicating that phosphorylated APE-1 is inactive as an endonuclease (44), we examined whether the HOCl-induced increase in the level of APE-1 polypeptide is accompanied by an increase in endonuclease activity. Extracts from HOCl-treated HeLa cells showed more than a 2-fold increase in APE activity relative to extracts from mock-treated cells (Fig. 5). A similar increase in the level of APE-1 polypeptide was observed in this particular experiment (Fig. 5 Inset). Because APE-1 activity appears to be limiting in the repair of ROS-induced DNA strand breaks by HeLa whole cell extract (28), it is likely that an increase in APE-1 activity would enhance the BER process in vivo.
Figure 5.
Increased APE activity after HOCl treatment. HeLa cells were treated with HOCl (130 nM), and APE-1 levels in mock- and HOCl-treated lysates were assessed by Western blot analysis (Inset). Aliquots of 10 μg total protein of nontreated (a) and HOCl-treated (b) cells were used. APE activity was assayed in extracts of treated (•) and untreated control (○) cells.
APE-1 Mediates Cellular Resistance to Toxic Doses of ROS.
We, therefore, asked whether increases in the level and activity of APE-1, and its accumulation in the nucleus, lead to increased in cellular resistance to toxic doses of ROS. HeLa S3 cells were treated with 130 nM HOCl, and 12 h later (when APE-1 activation was found to be the maximum), aliquots of treated and control cells were exposed to various doses of several genotoxic agents, namely H2O2, bleomycin, and MMS, γ-rays, UVC light (254 nm), and HOCl. The dose range for each agent was chosen on the basis of published data and prior trial experiments. Cell survival was quantitated by clonogenic assays as described in Materials and Methods. Fig. 6A shows a significant increase in resistance of HOCl-treated cells (relative to the control) for H2O2, bleomycin, and MMS. Thus, for example, with 0.2 mM MMS, which killed 80% of the control cells, 70% of the treated cells survived. A small increase in γ-ray resistance was also observed for the treated cells. However, no significant effect on the sensitivity to HOCl and UVC light was observed as a result of pretreatment with HOCl. Similar increases in resistance to H2O2, bleomycin, and MMS after HOCl pretreatment are similarly observed in the Chinese hamster V79 lung cell line, and they are accompanied by a similar increase in APE-1 polypeptide level. More remarkably, the cell killing profile as a function of toxicant concentration was nearly identical for HeLa and V79 cells (data not shown). We also observed that the enhanced resistance of HOCl-treated cells was absent when the cells were treated with H2O2 or MMS 24 (Fig. 6B) to 36 h (data not shown) after treatment with HOCl. This observation supports our conclusion that increased resistance was due to activation of APE-1 that occurs transiently and is absent 24 to 36 h after HOCl treatment, when APE-1 went back down to the basal level.
Activation of APE-1 by Other ROS.
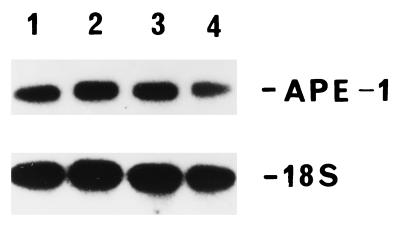
It is important to test whether APE-1 activation is unique to HOCl, or whether other ROS are also capable of eliciting this response. Nontoxic or slightly toxic doses of bleomycin (5 μg/ml), phenazine methosulfate (33) (5 μM), and H2O2 (50 μM) were also found to activate APE-1 at the polypeptide (Fig. 7) and RNA levels (data not shown). Ionizing radiation also increased the APE-1 mRNA level in a dose-dependent manner in HeLa cells (Fig. 1B). The extent and kinetics of APE-1 induction after irradiation with 2 Gy (a slightly toxic dose) were different from those observed with HOCl (Fig. 1 Ab and Bb).
Figure 7.
Induction of APE-1 by various ROS. HeLa cells were mock-treated (lane A) or treated with 5 μg/ml bleomycin (lane B), 5 μM phenazine methosulfate (lane C), or 50 μM H2O2 (lane D). At the indicated times, aliquots of cells were harvested, and Western blot analysis for APE-1 level was carried out with 10 μg of protein from each lysate.
Activation of APE-1 Is ROS Specific.
It was somewhat surprising that the APE-1 gene was not activated by either alkylating agents [2 mM MMS or 50 μM N,N′-bis(2-chloroethyl)-N-nitrosourea] or 10 J/m2 UVC light (Fig. 8). Activation of APE-1 exclusively by ROS and ROS generators sets it apart from most other DNA damage-inducible genes, which show similar responses to a variety of genotoxic agents (45–47).
Figure 8.
Lack of APE-1 activation by alkylating agents and UV light. Logarithmic-phase HeLa cells were mock-treated (lane 1), treated with 2 mM MMS (lane 2) or 50 μM N,N′-bis(2-chloroethyl)-N-nitrosourea (lane 3), or exposed to 254-nm UV light at 10 J/m2 (lane 4). Cells were incubated further for 9 h and harvested for Northern blot analysis. The blot was reprobed with 18S ribosomal RNA (Lower).
DISCUSSION
An “adaptive response” was first observed by Cairns and his collaborators in E. coli, which, after treatment with a sublethal level of an alkylating agent, developed transient resistance to a subsequent toxic dose of alkylating agents (29). This adaptive response was later found to result from activation of a set of genes whose products were involved in the repair of genotoxic and mutagenic DNA damage induced by alkylating agents (reviewed in ref. 30). Subsequently, an adaptive response of E. coli to ROS was observed, and it was found to involve activation of not only APE (endonuclease IV) but also other genes whose products counter the deleterious effects of ROS (17).
Several examples of adaptive responses of mammalian cells to ROS have been reported (32, 34), but the responses have not been directly linked to enhanced repair of ROS-induced genotoxic damage. Our report documents that an adaptive response to ROS, observed in human cells, fulfills the original definition of Cairns and his collaborators (29). We have shown that ROS transiently activate APE-1, and that cells with elevated APE-1 develop resistance to genotoxic ROS and ROS generators. Although we have not yet directly proved causality, our recent studies (28) and another published report (48) strongly suggest that APE-1 is essential in the repair of ROS-induced cytotoxic DNA damage.
Our results on the selective resistance of adapted cells to the various agents (Fig. 6) can be explained as follows. Assuming that an increase in the APE-1 level enhances BER, increased resistance of the cells, after adaptation treatment, to H2O2, bleomycin, and MMS should be due to increased repair of toxic AP sites and DNA strand break lesions via the BER pathway. UVC induces pyrimidine photoproducts in DNA, whose repair via the nucleotide excision repair (NER) pathway does not involve APE-1. Thus APE-1 activation did not affect the UV sensitivity of HOCl-treated cells. We can infer at the same time that ROS did not activate the NER pathway. Although γ-rays activated APE-1, we observed that the increase in APE-1 had a modest effect on γ-ray sensitivity. Because cell killing by ionizing radiation is mostly due to DNA double-strand breaks, whose repair does not involve BER and APE, the low level of observed protection with a high dose of γ-rays could reflect the small contribution of single-strand breaks to cell killing. The lack of protection of HOCl-activated cells against HOCl itself can be explained by the possibility that HOCl, a powerful oxidant that reacts with primary amino groups, is more likely to kill cells by membrane oxidation than by DNA damage (1). Finally, the resistance of adapted cells to MMS, which did not itself activate APE-1, may be a gratuitous response in which enhanced BER activity increased repair of DNA strand breaks during MMS damage repair.
The ROS-induced APE-1 activation is not limited to the human cells. Although there is one report on the absence of such activation (49), several other recent studies confirm our observation that ROS induce APE-1 in human and other mammalian cells (B. Kaina, S. A. Leadon, and B. T. Mossman, personal communication). Furthermore, HOCl, generated in and released from neutrophils during the inflammatory response, causes strong oxidative stress in vivo (50). Thus, in vitro activation of APE-1 by sublethal levels of a wide variety of ROS and ROS generators provides a strong argument in support of the in vivo occurrence of such activation.
That activation occurs at the level of transcription is not surprising. However, several unusual features of APE-1 activation are evident. The late activation occurring 6–9 h after ROS exposure is unexpected, and it suggests involvement of multiple signaling intermediates. Furthermore, migration of APE-1 to the nucleus in ROS-treated cells suggests additional, uncharacterized features of this enzyme. It was recently shown that APE-1 could be phosphorylated at multiple Ser/Thr residues by casein kinases and protein kinase C, and that phosphorylation of Thr-233 might be responsible for inactivation of the endonuclease activity (44). It thus appears that, in addition to regulation at the transcriptional level, the function and activity of APE could be modulated by posttranslational modification.
Activation of APE-1 may be somewhat unexpected because it is a relatively abundant protein. However, as pointed out earlier, because of the rather low intrinsic 3′ DNA phosphoesterase activity of the protein, it may still be limiting in the multiprotein repair pathway, and a change in its level will affect overall repair (27, 28).
The molecular mechanisms of transcriptional activation of the APE-1 gene remain to be elucidated. Although APE-1 modulates DNA binding activity of p53 (51), APE-1 activation itself may not involve p53 because it occurs in primary fibroblasts with wild-type p53 as well as in HeLa cells which are phenotypically deficient in the protein (52). The fact that APE-1 is activated exclusively by ROS is intriguing. The transcription factors NF-κB, C/EBP, AP-1, and Egr-1 have been implicated in activation of several irradiation- and ROS-responsive genes, although many of these genes are also activated by a variety of genotoxic agents (8, 47, 53). However, the human APE-1 promoter contains consensus sequences for binding NF-κB but not functional AP-1 (49, 54). Furthermore, the lack of APE-1 activation by phorbol ester, which activates protein kinase C, argues against involvement of AP-1. On the other hand, that H7 and staurosporine, inhibitors of protein kinase C, prevented APE-1 activation suggests involvement of the kinase in the process (Fig. 3B). However, because such inhibitors are somewhat promiscuous, other serine/threonine kinases may well be involved in ROS-mediated cellular signaling. ROS, including O2⨪, H2O2, ⋅OH, HOCl, and peroxyl radicals, have been shown to be involved in various biological processes, including cell growth and apoptosis, presumably by altering intracellular Ca2+ level, and thereby affecting Ca2+-dependent kinases, phosphatases, and DNA binding activity of transcription factors (6). hAPE-1 has been implicated in Ca2+-dependent negative regulation of genes, including its own (42, 54), so it is possible that ROS activation of APE-1 is indirect and is mediated by Ca2+-dependent signaling processes.
Finally, because APE-1 arguably plays a central role in various cellular activities, including gene regulation and DNA repair, it is important to address in vivo significance of activation of the hAPE-1 gene, in regard to the adaptive response and cellular homeostasis.
Acknowledgments
We thank Dr. Pricscilla K. Cooper of Lawrence Berkeley Laboratories for a critical reading of the manuscript and for important suggestions, and Dr. D. Konkel for carefully editing the manuscript. Dr. S. Seki’s gift of hAPE-1 plasmid DNA is also appreciated. We also acknowledge the valuable technical help of Ms. Julie Lock and secretarial assistance of Ms. Wanda Smith. This research was supported by U.S. Public Health Service Grants ES08457, CA53791, and AG10514 and by National Institute on Environmental Health Sciences Center Grant ES06676.
ABBREVIATIONS
- AP
apurinic/apyrimidinic
- APE
AP-endonuclease
- hAPE-1
human APE 1
- BER
base excision repair
- ROS
reactive oxygen species
- MMS
methyl methanesulfonate
References
- 1.Grisham M B, McCord J M. In: Physiology of Oxygen Radicals. Taylor A E, Matalon S, Ward P A, editors. Baltimore: Waverly; 1986. pp. 1–18. [Google Scholar]
- 2.Gotz M E, Kunig G, Riederer P, Youdim M B. Pharmacol Ther. 1994;63:37–122. doi: 10.1016/0163-7258(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 3.Parkins C S, Dennis M F, Stratford M R, Hill S A, Chaplin D J. Cancer Res. 1995;55:6026–6029. [PubMed] [Google Scholar]
- 4.Ward J F. Int J Radiat Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 5.Tyrrell R M, Keyse S M. J Photochem Photobiol B. 1990;4:349–361. doi: 10.1016/1011-1344(90)85014-n. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki Y J, Forman H J, Sevarian A. Free Radical Biol Med. 1997;22:269–285. doi: 10.1016/s0891-5849(96)00275-4. [DOI] [PubMed] [Google Scholar]
- 7.Ames B N. Science. 1983;221:1256–1264. doi: 10.1126/science.6351251. [DOI] [PubMed] [Google Scholar]
- 8.Papaconstantinou J. Ann N Y Acad Sci. 1994;719:195–211. doi: 10.1111/j.1749-6632.1994.tb56829.x. [DOI] [PubMed] [Google Scholar]
- 9.Harman D. Mutat Res. 1992;275:257–266. doi: 10.1016/0921-8734(92)90030-s. [DOI] [PubMed] [Google Scholar]
- 10.Breen A P, Murphy J A. Free Radical Biol Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 11.Dedon P C, Goldberg I H. Chem Res Toxicol. 1992;5:311–332. doi: 10.1021/tx00027a001. [DOI] [PubMed] [Google Scholar]
- 12.Loeb L A, Preston B D. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 13.Wallace D C. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 14.Doetsch P W, Cunningham R P. Mutat Res. 1990;236:173–201. doi: 10.1016/0921-8777(90)90004-o. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg E C, Walker G C, Seide W. DNA Repair. San Francisco: Freeman; 1994. pp. 135–190. [Google Scholar]
- 16.Kane C M, Linn S. J Biol Chem. 1981;256:3405–3414. [PubMed] [Google Scholar]
- 17.Demple B, Harrison L. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 18.Demple B. Annu Rev Genet. 1991;25:315–337. doi: 10.1146/annurev.ge.25.120191.001531. [DOI] [PubMed] [Google Scholar]
- 19.Seki S, Hatsushika M, Watanabe S, Akiyama K, Nagao K, Tsutsui K. Biochim Biophys Acta. 1992;1131:287–299. doi: 10.1016/0167-4781(92)90027-w. [DOI] [PubMed] [Google Scholar]
- 20.Radicella J P, Dherin C, Desmaze C, Fox M S, Boiteux S. Proc Natl Acad Sci USA. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roldan-Arjona T, Wei Y F, Carter K C, Klungland A, Anselmino C, Wang R P, Augustus M, Lindahl T. Proc Natl Acad Sci USA. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenquist T A, Zharkov D O, Grollman A P. Proc Natl Acad Sci USA. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu R, Nash H M, Verdine G L. Curr Biol. 1997;7:397–407. doi: 10.1016/s0960-9822(06)00187-4. [DOI] [PubMed] [Google Scholar]
- 24.Hilbert T P, Chaung W, Boorstein R J, Cunningham R P, Teebor G W. J Biol Chem. 1997;272:6733–6740. doi: 10.1074/jbc.272.10.6733. [DOI] [PubMed] [Google Scholar]
- 25.Aspinwall R, Rothwell D G, Roldan-Arjona T, Anselmino C, Ward C J, Cheadle J P, Sampson J R, Lindahl T, Harris P C, Hickson I D. Proc Natl Acad Sci USA. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitra S, Hazra T K, Roy R, Ikeda S, Biswas T, Lock J, Boldogh I, Izumi T. Mol Cells. 1997;7:305–312. [PubMed] [Google Scholar]
- 27.Suh D, Wilson D M, III, Povirk L F. Nucleic Acids Res. 1997;25:2495–2500. doi: 10.1093/nar/25.12.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izumi T, Boldogh I, Ramana C V, Hsieh C C, Saito H, Papaconstantinou J, Mitra S. In: Repair of Oxidative DNA Damage and Aging. Dizdaroglu M, editor. New York: Plenum; 1998. , in press. [Google Scholar]
- 29.Cairns J, Robins P, Sedgwick B, Talmud P. Prog Nucleic Acid Res Mol Biol. 1981;26:237–244. doi: 10.1016/s0079-6603(08)60408-0. [DOI] [PubMed] [Google Scholar]
- 30.Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- 31.Demple B, Halbrook J. Nature (London) 1983;304:466–468. doi: 10.1038/304466a0. [DOI] [PubMed] [Google Scholar]
- 32.Wolff S, Jostes R, Cross F T, Hui T E, Afzal V, Wiencke J K. Mutat Res. 1991;250:299–306. doi: 10.1016/0027-5107(91)90185-q. [DOI] [PubMed] [Google Scholar]
- 33.Marsh J P, Mossman B T. Cancer Res. 1991;51:167–173. [PubMed] [Google Scholar]
- 34.Vile G F, Basu-Modak S, Waltner C, Tyrrell R M. Proc Natl Acad Sci USA. 1994;91:2607–2610. doi: 10.1073/pnas.91.7.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manley J L, Fire A, Samuels M, Sharp P A. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 36.Izumi T, Mitra S. Carcinogenesis. 1998;19:525–527. doi: 10.1093/carcin/19.3.525. [DOI] [PubMed] [Google Scholar]
- 37.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Greene & Wiley; 1990. [Google Scholar]
- 38.Boldogh I, AbuBakar S, Deng C Z, Albrecht T. J Virol. 1991;65:1568–1571. doi: 10.1128/jvi.65.3.1568-1571.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Flora S, Cesarone C F, Balansky R M, Albini A, D’Agostini F, Bennicelli C, Bagnasco M, Camoirano A, Scatolini L, Rovida A. J Cell Biochem Suppl. 1995;22:33–41. doi: 10.1002/jcb.240590806. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, Moore D H, Broshears J, Liu L F, Wilson T M, Kelley M R. Anticancer Res. 1997;17:3713–3719. [PubMed] [Google Scholar]
- 41.Duguid J R, Eble J N, Wilson T M, Kelley M R. Cancer Res. 1995;55:6097–6102. [PubMed] [Google Scholar]
- 42.Okazaki T, Chung U, Nishishita T, Ebisu S, Usuda S, Mishiro S, Xanthoudakis S, Igarashi T, Ogata E. J Biol Chem. 1994;269:27855–27862. [PubMed] [Google Scholar]
- 43.Xanthoudakis S, Curran T. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yacoub A, Kelley M R, Deutsch W A. Cancer Res. 1997;57:5457–5459. [PubMed] [Google Scholar]
- 45.Fornace A J., Jr Annu Rev Genet. 1992;26:507–526. doi: 10.1146/annurev.ge.26.120192.002451. [DOI] [PubMed] [Google Scholar]
- 46.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. Nature (London) 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 47.Schreck R, Albermann K, Baeuerle P A. Free Radical Res Commun. 1992;17:221–237. doi: 10.3109/10715769209079515. [DOI] [PubMed] [Google Scholar]
- 48.Ono Y, Furuta T, Ohmoto T, Akiyama K, Seki S. Mutat Res. 1994;315:55–63. doi: 10.1016/0921-8777(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 49.Harrison L, Ascione A G, Wilson D M, 3rd, Demple B. J Biol Chem. 1995;270:5556–5564. doi: 10.1074/jbc.270.10.5556. [DOI] [PubMed] [Google Scholar]
- 50.Thelen M, Dewald B, Baggiolini M. Physiol Rev. 1993;73:797–821. doi: 10.1152/physrev.1993.73.4.797. [DOI] [PubMed] [Google Scholar]
- 51.Jayaraman L, Murthy K G, Zhu C, Curran T, Xanthoudakis S, Prives C. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 52.Isaacs J S, Chen P C, Garza A, Hansen M F, Barrett J C, Weissman B, E. Oncogene. 1997;14:1669–1678. doi: 10.1038/sj.onc.1201001. [DOI] [PubMed] [Google Scholar]
- 53.Hallahan D E, Gius D, Kuchibhotla J, Sukhatme V, Kufe D W, Weichselbaum R R. J Biol Chem. 1993;268:4903–4907. [PubMed] [Google Scholar]
- 54.Izumi T, Henner W D, Mitra S. Biochemistry. 1996;35:14679–14683. doi: 10.1021/bi961995u. [DOI] [PubMed] [Google Scholar]