Abstract
The establishment of differential gene expression in sporulating Bacillus subtilis involves four protein components, one of which, SpoIIAA, undergoes phosphorylation and dephosphorylation. We have used NMR spectroscopy to determine the solution structure of the nonphosphorylated form of SpoIIAA. The structure shows a fold consisting of a four-stranded β-sheet and four α-helices. Knowledge of the structure helps to account for the phenotype of several strains of B. subtilis that carry known spoIIAA mutations and should facilitate investigations of the conformational consequences of phosphorylation.
The protein SpoIIAA participates, via phosphorylation and dephosphorylation, in the four-component system that regulates the sporulation sigma factor σF. Sporulation is a response of the soil bacterium Bacillus subtilis to nutrient deprivation. Instead of continuing normal vegetative cell division, the bacterium divides asymmetrically, and the resulting two-chamber sporangium enters a pathway of differential gene expression that leads to the formation of a dormant cell type called the endospore (1, 2). Differential gene expression depends on specialized transcription factors called sigma factors that direct the RNA polymerase to transcribe specific genes in one or other of the two chambers at various stages of sporulation. The first sporulation-specific sigma factor to be activated is σF; transcription that depends on σF is essential for the remaining sigma factors to become active in turn (3–6). Early in sporulation, SpoIIAA is in the phosphorylated state (SpoIIAA-P) (7), as a result of the activity of the ATP-dependent protein kinase SpoIIAB (8). SpoIIAA-P has very low affinity for SpoIIAB (9). About 80 min after the initiation of sporulation a specific phosphatase, SpoIIE (7, 10, 11), begins to hydrolyse SpoIIAA-P, and the resulting SpoIIAA again becomes a substrate for SpoIIAB. SpoIIAB is also an anti-sigma factor that in its free form inhibits σF by binding to it (8, 12). Competition by SpoIIAA (the anti-anti-sigma factor) for binding to SpoIIAB releases σF activity (9, 13–15). We now report the structure of the nonphosphorylated form of SpoIIAA, as an initial step toward elucidating the structural basis of the regulation of differential gene expression in this organism.
MATERIALS AND METHODS
Production of the SpoIIAA Samples.
SpoIIAA from B. subtilis was overproduced in Escherichia coli by using the T7 RNA polymerase system (16). Details of the overproduction and purification were as described (13), except that the final gel filtration was over Sephadex G-50 equilibrated in NMR sample buffer (25 mM NaCl/25 mM K2HPO4/1 mM DTT, pH 7.0). N-terminal sequencing showed that the protein was at least 95% pure and that Met-1 had been cleaved posttranslationally to yield a protein with a N-terminal serine (A.C. Willis and M.D.Y., unpublished results). 2H2O was added to the samples at 5% (vol/vol) and the pH was adjusted to 6.4. Uniform 13C and 15N isotope labeling was accomplished by growing the cells on minimal medium containing 15NH4Cl and [13C]glucose as sole nitrogen and carbon sources, which yielded 10–15 mg of protein per liter.
NMR Spectroscopy.
The NMR experiments were performed on samples containing ∼1 mM unlabeled or 0.7 mM uniformly 15N-labeled and 0.7 mM uniformly 13C- and 15N-labeled SpoIIAA protein at 25°C. The triple resonance experiments were performed at 500 MHz, and the spectra for the collection of conformational constraints were recorded on a home-built 750-MHz spectrometer. Sequential assignment was obtained by analysis of sensitivity-enhanced (17) HNCA,¶ CBCA(CO)NH,¶ HBHA(CBCACO)NH,¶ and 15N-correlated nuclear Overhauser effect spectroscopy (NOESY) spectra. Distance constraints were collected from NOESY spectra in H2O and 2H2O of the unlabeled sample and a 15N-correlated NOESY spectrum. The mixing time was set to 125 ms. The 3JHNα coupling constants for dihedral angle constraints were determined from a heteronuclear multiple quantum coherence J coupling (18) spectrum of the 15N-labeled sample. Spectral processing was done with felix software (felix 2.3, Biosym Technologies, San Diego), and the program xeasy (19) was used for the spectral analysis.
Collection of Conformational Constraints.
The nuclear Overhauser effect (NOE) intensities were calibrated automatically against the corresponding upper distance bounds with the auxiliary program caliba in dyana.‖ The vicinal scalar coupling constants 3JHNα were determined by curve fitting to traces extracted from the heteronuclear multiple quantum coherence J coupling spectrum. The allowed intervals for the dihedral angles φ, ψ, and χ1, which were compatible with the intraresidual and sequential backbone distance constraints and the coupling constants, were defined by the auxiliary program habas (20) in dyana.‖ In addition, based on the identification of a number of backbone amide protons in the 2H2O-NOESY spectrum as slowly exchanging, hydrogen bonds were implemented in the regular secondary structure at a late stage in the structure determination.
Determination and Analysis of the Three-Dimensional Structure.
Structure calculations were performed with the torsion angle dynamics software dyana.‖ The total number of steps was set to 6,000, the high-temperature stage constituting, by default, one-fifth of them. In the end, conjugent gradient minimization with 1,000 steps was performed. The final 24 structures of 100 were selected based on their agreement with the experimental data (low values of the target function, NOE, and torsion angle violations; see Table 1). For the visual display and quantitative analysis of the calculated structures, the molecular graphics software molmol (22) was used. Global superposition was made for the backbone atoms N, Cα, and C′ of residues 3–72 and 77–113, and the mean coordinates, 〈NMR〉, were obtained by averaging the Cartesian coordinates of each atom after the superposition. The rms deviations in Table 1 are reported both for the backbone atoms (N, Cα, and C′) and for all heavy atoms after the superposition of the indicated region. The quality of the calculated structures was assessed by the program procheck (21). Structural homologues were searched for by the Internet server dali 2.0 (23), which scans a representative database of the three-dimensional folds in Protein Data Bank.
Table 1.
Summary of the experimental constraints and the structure calculation
| Value | |
|---|---|
| Conformational constraints | |
| NOE-based distance bounds | |
| Intraresidue, |j–i| = 0 | 876 |
| Sequential, |j–i| = 1 | 578 |
| Medium range, 2 ≤ |j–i| ≤ 4 | 286 |
| Long-range, |j–i| ≥ 5 | 328 |
| Total | 2,068 |
| Dihedral angle constraints | |
| 3JHNα and local NOE data based* backbone φ and ψ | 208 |
| 3JHNα and local NOE data based* χ1 | 81 |
| 13Cα chemical shift based backbone φ and ψ | 128 |
| Hydrogen-bond constraints | 45 |
| Quality of the calculated structures | |
| Violations† | |
| Distance bounds (>0.7 Å) | 4 |
| Twenty-four best structures 〈NMR〉§ | 3 |
| Dihedral angle constraints (>20°) | 0 |
| Twenty-four best structures‡ 〈NMR〉§ | 1 |
| Target function, average ± SD (range) | 43.9 ± 2.2 (39.6–50.6) |
| Ramachandran analysis¶ | |
| Residues in the most favored regions, % | 66.3 |
| Additional allowed regions, % | 24.8 |
| Generously allowed regions, % | 6.9 |
| Disallowed regions¶, % | 2.0 |
| rms deviation differences**, Å | |
| Residues | Backbone N, Cα, and C′ All heavy atoms |
| 2–117 | 0.62 ± 0.121.00 ± 0.11 |
| 3–72, 77–113 | 0.52 ± 0.100.96 ± 0.10 |
| I helix (Thr-26–Leu-38) | 0.26 ± 0.100.74 ± 0.15 |
| II helix (Ser-58–Lys-72) | 0.42 ± 0.121.04 ± 0.21 |
| III helix (Lys-88–Ser-94) | 0.08 ± 0.050.67 ± 0.08 |
| IV helix (Glu-106–Thr-112) | 0.11 ± 0.060.67 ± 0.11 |
| I β-strand (Gly-4–Lys-10) | 0.11 ± 0.070.48 ± 0.14 |
| II β-strand (Val-13–Thr-19) | 0.10 ± 0.040.59 ± 0.18 |
| III β-strand (His-45–Asn-49) | 0.05 ± 0.020.54 ± 0.09 |
| IV β-strand (Glu-77–Cys-81) | 0.11 ± 0.030.80 ± 0.27 |
Obtained from the program habas (20).
The distance constraints that are violated in the bundle of 24 best structures are L3 Qβ to T26 Hα, H45 Hɛ1 to G76 Hα2, H45 Hɛ1 to G76 Qα, and D92 Hα to F97 Hζ. The distance constraints that are violated in the mean structure are L3 Qβ to T26 Hα, H45 Hɛ1 to G76 Qα, and D92 Hα to F97 Hζ. The dihedral angle constraint that is violated in the mean structure is L50 χ1.
Violated in 12 or more structures.
Mean structure was obtained by averaging the coordinates of the 24 best structures after superposition of the N, Cα, C′ backbone atoms in the structured regions containing residues 3–72 and 77–113.
Determined by the program procheck (21).
Residues with φ and ψ dihedral angles that lie in the unfavorable regions are Gly-20, Gly-76, Glu-103, Ser-105, and Ser-116. All of these residues are found in loops and not in the secondary structural elements.
rms deviation of the 24 best structures vs. the mean structure.
The 1H, 15N, and 13C resonance assignments observed for SpoIIAA, the angle bar plot of φ and ψ dihedrals in the best structures, plots of NOEs and rms deviation per residue, and more details about the materials and methods used are available at http://www.ocms.ox.ac.uk/~comfort/spoiiaa_supplement.html.
RESULTS AND DISCUSSION
Structure of SpoIIAA.
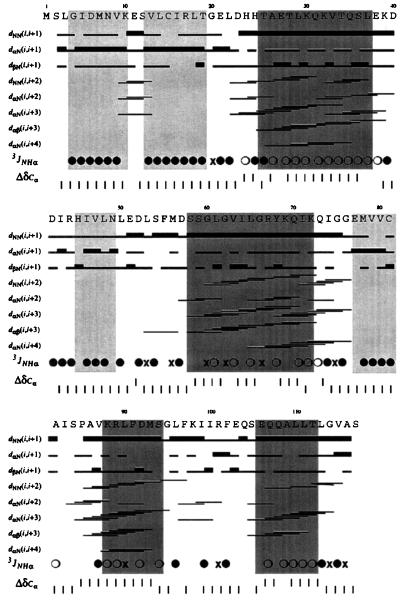
We have determined the structure of the nonphosphorylated form of SpoIIAA in solution by NMR spectroscopy by using unlabeled, 15N-labeled, and 15N- and 13C-labeled samples. The sequential assignment of backbone resonances, which was obtained by means of triple resonance experiments, is complete. A nearly complete side-chain assignment (84%) was obtained through analysis of the NOESY spectra. A summary of the experimental constraints used in the structure determination is presented in Table 1. The total number of nonredundant NOE-based distance restraints is 2,068 and the number of 3JHNα coupling constants is 81. The presence of four α-helices and four β-strands in the sequence can readily be established by inspection of the experimental data, as illustrated (Fig. 1). The patterns of characteristic NOE connectivities, values of the 3JHNα coupling constants, and deviation from the random coil values of the 13Cα chemical shift are in accord with the location of the secondary-structure elements (Fig. 1). The structure calculation method was simulated annealing in torsion angle space.
Figure 1.
Summary of the 3JNHα coupling constants and sequential- and medium-range NOE connectivities for backbone atoms and secondary-structure elements. •, 3JNHα coupling constant >8 Hz; ○, 3JNHα <6 HZ; ×, 6 Hz < 3JNHα < 8 Hz. The plot distinguishes between weak and strong (thin and thick bar) only for sequential dαN, dNN, and dβN distance constraints, the cutoff of the strong constraint being 3.0, 3.6, and 3.6 Å, respectively. The normalized deviations of the chemical shift of the 13Cα atoms from their random coil values (24) are shown on the bottom line. Light shading, β-strand; dark shading, α-helical region.
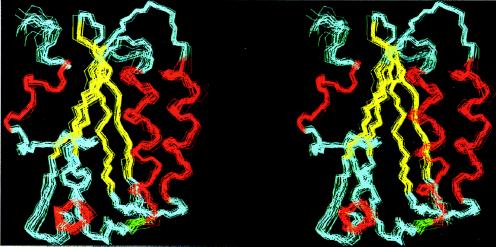
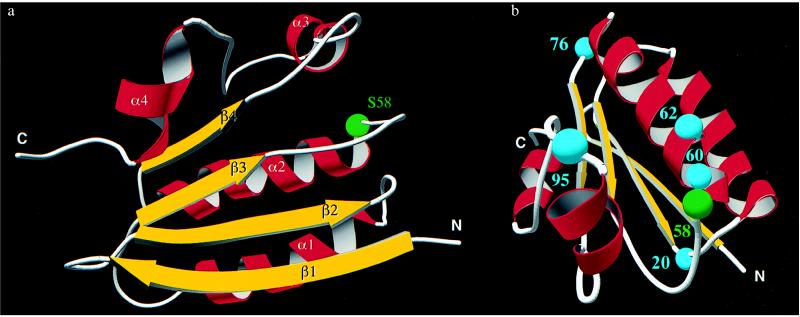
The 25°C solution structure of SpoIIAA is represented by a set of 24 energy-minimized conformers after a pairwise backbone superposition of the well-defined regions to the mean structure, as shown in Fig. 2. The quality of the structure determination is indicated by the average rms deviation of the individual conformers from their mean (〈NMR〉), which is 0.62 Å for backbone atoms and 1.00 Å for all heavy atoms (Table 1), after global superposition of all backbone atoms. The main secondary-structure elements, that is, the four α-helices and four β-strands, are well-defined. The tertiary fold consists of a four-stranded β-sheet, two α-helices above it, one short α-helix at the side, and a short C-terminal α-helix, as illustrated (Fig. 3a).
Figure 2.
Stereo view of the superposition of the backbone atoms N, Cα, and C′ to the mean in the structured regions (residues 3–72 and 77–113) of the 24 best conformers. The α-helices are shown in red, the β-strands are in yellow, and coils are in cyan.
Figure 3.
(a) Global fold of the nonphosphorylated SpoIIAA. The position of the phosphorylatable Ser-58 is highlighted by a green sphere. The figure is drawn by using molscript (25), povray (http:/www.povray.org/), and povscript (http:/www.rose.brandeis.edu/users/peisach/rayscript/). (b) Mutational sites in SpoIIAA. Gly-20, Gly-60, Gly-62, Gly-76, and Gly-95 are denoted with cyan spheres. The figure is drawn by using molscript, povray, and povscript.
The first helix (Thr-26–Leu-38) displays a characteristic N-capping motif (26) and a regular pattern of hydrophobic residues in every fourth position, Leu-30, Val-34, Leu-38, interspersed with charged or polar residues. This renders the helix amphipathic. The second α-helix comprising residues Ser-58–Lys-72 also has amphipathic character. The third helix (Lys-88–Ser-94) is very well-defined as practically all the characteristic helical dNN, dαN, and dαβ NOE connectivities are present (Fig. 1). The position of the third α-helix, however, is less well defined with respect to the rest of the protein structure in the present calculation. The aromatic rings of the residues Phe-91, Phe-97, and Phe-102 anchor this protruding region by inserting into the hydrophobic core. The fourth α-helix, composed of residues Glu-106–Thr-112, is short and well-defined.
The two β-strands at the beginning of the sequence, containing residues Gly-4–Lys-10 and Val-13–Thr-19, align in an antiparallel fashion. They are connected by three characteristic cross-strand Hα–Hα and three cross-strand HN–HN constraints. There is a well-defined β-hairpin (tight turn of type I) between the two β-strands, from position 10 to position 13. The third β-strand (His-45–Asn-49) aligns with residues Val-13–Arg-17 in the second β-strand in a parallel fashion. It is flanked on its other side by a short parallel β-strand containing residues Glu-77–Cys-81. In this region, the characteristic cross-strand dαN, dNα, and dNN connectivities are too few to give rise to regular β-structure, although they are confirmed by a high number of side-chain connectivities. The spectral analysis of the residues in this region is complicated by overlap. The absence of medium- or long-range NOEs for residues Glu-51–Asp-57 (Fig. 1), indicates that this is a loop region. In addition, the region containing residues 73–76 and four C-terminal residues have fewer NOEs and are thus less well defined than other regions.
A search in the Protein Data Bank by dali 2.0 (23) did not detect structural homologues (z score < 5.0). However, A. Murzin (personal communication) has pointed out that in the recently published structure of phosphatidylinositol transfer protein from yeast (27), the core of the C-terminal domain shows a certain topological similarity to the fold of SpoIIAA.
The Phosphorylation Site.
The phosphorylation site in SpoIIAA is Ser-58, which is at the beginning of the second α-helix. The N terminus of an α-helix is a favorable location for a phospho group (28) because it is stabilized by the partial positive charge of the helix dipole and by interactions with the exposed backbone NH groups. Ser-58 is also close to the N-terminal end of the first α-helix, the closest backbone distance being around 5 Å in the present structure. The proximity of this helix dipole is also presumably a stabilizing factor, provided that no large conformational changes take place upon phosphorylation of SpoIIAA. The dissociation constant of the complex consisting of two monomers of dephosphorylated SpoIIAA per SpoIIAB dimer and containing ADP, is in the region of 50 nM (14, 15). It has been proposed that phosphorylation of SpoIIAA prompts a major conformational change in the protein architecture, preventing it from binding to SpoIIAB (29). Alternatively, the phospho group itself may sterically interfere with complex formation even though it imposes only minor conformational adjustments in its immediate environment. We are now studying the structure of phosphorylated SpoIIAA to distinguish between these possibilities.
Possible Effects of Known Mutations.
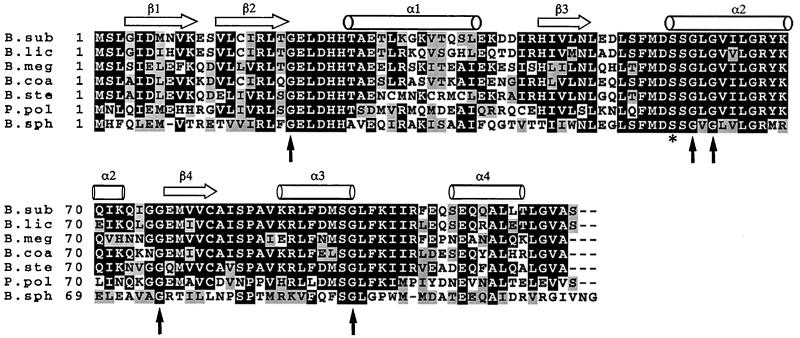
Diederich et al. (13) have shown that mutation of Ser-58 to Asp blocks σF activity in vivo but the mutation of Ser-58 to Ala triggers release of σF prematurely. It has also been shown that the mutations G60R and G62D abolish function but S58N leaves SpoIIAA partially functional (30) (see Fig. 3b). These results can be readily explained in terms of modifications in and around the phosphorylation site. Furthermore, the new structure shows that the β2–α1 loop and the beginning of the α3-helix are also in the vicinity of the phosphorylation site. We note that these regions, with sequences GELDHH and VKRL, respectively, are highly conserved in the known sequences of the SpoIIAA from six species of Bacillus as well as that from Paenibacillus polymyxa (Fig. 4). Because inspection of the structure shows no obvious structural requirement for conservation in this region, this observation suggests that the β2–α1 loop may be involved in some way in the function of SpoIIAA—a proposal that is supported by the observation that the double mutant G20S, S58N is noticeably different in phenotype from the single mutant S58N (30). [Certain sporulation-specific proteins whose synthesis depends on the activity of σF are produced to a perceptible extent in the mutant S58N but not in G20S, S58N (30).] Two further mutations in SpoIIAA are known: G76E blocks activity and G95D leaves partial function (30). The G76 mutation is in the α2–β4 loop at the opposite end of the molecule. This glycine and its glycine neighbor are, however, in a relatively tight turn and the change to a glutamic acid might cause a global structural change; we note that all the SpoIIAA homologues have glycine at this position (Fig. 4). The residue Gly-95 is at the end of the α3-helix; it is possible that this region of the protein is at or near the SpoIIAB binding site. From these considerations, it seems likely that the SpoIIAA/SpoIIAB interaction site involves the β3–α2 loop (containing the phosphorylation site), the β2–α1 loop, and the α3-helix, but the possibility that one or more of these regions of SpoIIAA is involved instead (or as well) in an interaction with SpoIIE is not excluded by our results.
Figure 4.
Multiple sequence alignment of SpoIIAA, obtained through a search in the EMBL/GenBank DNA data base with fasta version 3.0.7 (31) using the program default parameters. The amino acids are denoted by single-letter code, dashes indicate gaps, and amino acid position is indicated on the left. Consensus residues that are present in at least four of seven sequences are boxed. Conserved changes are indicated by stippled boxes. The location of the β-strands is marked with strands and the α-helical regions are marked by cylinders. The conserved phosphorylation site is marked with a star, and the sites of mutation (30) shown in Fig. 3b and discussed in the text are marked with vertical arrows. Accession numbers from the GenBank/EMBL and SwissProt data bases are as follows: SpoIIAA-B.sub, P10727; SpoIIAA-B.lic, P26777; SpoIIAA-B.meg, P35147; SpoIIAA-B.coa, Z54161; SpoIIAA-B.ste, L47358; SpoIIAA-P.pol, L47359; SpoIIAA-B.sph, L47360.
Acknowledgments
H.K. and the NMR facilities were funded through Oxford Centre for Molecular Sciences that is supported by the Biotechnology and Biological Sciences (BBSRC), the Medical (MRC) and the Engineering and Physical Sciences Research Councils (EPSRC) of the U.K. Support for D.C. was provided by James T. Comfort. Additional support was provided by BBSRC (M.L. and M.D.Y.) and the Wellcome Trust (I.D.C.).
ABBREVIATIONS
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1AUZ).
The nomenclature of these experiments is explained in ref. 17.
Güntert, P., Mumenthaler, C. & Wüthrich, K., XVIIth International Conference on Magnetic Resonance in Biological Systems, August 18–23, 1996, Keystone, CO.
References
- 1.Errington J. Trends Genet. 1996;12:31–34. doi: 10.1016/0168-9525(96)81386-2. [DOI] [PubMed] [Google Scholar]
- 2.Stragier P, Losick R. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 3.Losick R, Stragier P. Nature (London) 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 4.Karow M L, Glaser P, Piggot P J. Proc Natl Acad Sci USA. 1995;92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Londono-Vallejo J A, Stragier P. Genes Dev. 1995;9:503–508. doi: 10.1101/gad.9.4.503. [DOI] [PubMed] [Google Scholar]
- 6.Sun D, Stragier P, Setlow P. Genes Dev. 1989;3:141–149. doi: 10.1101/gad.3.2.141. [DOI] [PubMed] [Google Scholar]
- 7.Feucht A, Magnin T, Yudkin M D, Errington J. Genes Dev. 1996;10:794–803. doi: 10.1101/gad.10.7.794. [DOI] [PubMed] [Google Scholar]
- 8.Min K-T, Hilditch C M, Diederich B, Errington J, Yudkin M D. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 9.Alper S, Duncan L, Losick R. Cell. 1994;77:195–205. doi: 10.1016/0092-8674(94)90312-3. [DOI] [PubMed] [Google Scholar]
- 10.Arigoni F, Duncan L, Alper S, Losick R, Stragier P. Proc Natl Acad Sci USA. 1996;93:3238–3242. doi: 10.1073/pnas.93.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan L, Alper S, Arigoni F, Losick R, Stragier P. Science. 1995;270:641–644. doi: 10.1126/science.270.5236.641. [DOI] [PubMed] [Google Scholar]
- 12.Duncan L, Losick R. Proc Natl Acad Sci USA. 1993;90:2325–2329. doi: 10.1073/pnas.90.6.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diederich B, Wilkinson J F, Magnin T, Najafi S M A, Errington J, Yudkin M D. Genes Dev. 1994;8:2653–2663. doi: 10.1101/gad.8.21.2653. [DOI] [PubMed] [Google Scholar]
- 14.Duncan L, Alper S, Losick R. J Mol Biol. 1996;260:147–164. doi: 10.1006/jmbi.1996.0389. [DOI] [PubMed] [Google Scholar]
- 15.Magnin T, Lord M, Yudkin M D. J Bacteriol. 1997;179:3922–3927. doi: 10.1128/jb.179.12.3922-3927.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 17.Muhandiram D R, Kay L E. J Magn Res Series B. 1994;103:203–216. [Google Scholar]
- 18.Kay L E, Bax A. J Magn Reson. 1990;86:110–126. [Google Scholar]
- 19.Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K. J Biomol NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 20.Güntert P, Braun W, Billeter M, Wüthrich K. J Am Chem Soc. 1989;111:3997–4004. [Google Scholar]
- 21.Laskowski R A, MacArthur M V, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 22.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 24.Wishart D S, Sykes B D, Richards F M. J Mol Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 25.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 26.Richardson J S, Richardson D C. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 27.Sha B, Phillips S E, Bankaitis V A, Luo M. Nature (London) 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- 28.Hol W G J, van Duijnen P T, Berendsen H J C. Nature (London) 1978;273:443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- 29.Magnin T, Lord M, Errington J, Yudkin M D. Mol Microbiol. 1996;19:901–907. doi: 10.1046/j.1365-2958.1996.434964.x. [DOI] [PubMed] [Google Scholar]
- 30.Challoner-Courtney I J, Yudkin M. J Bacteriol. 1993;175:5636–5641. doi: 10.1128/jb.175.17.5636-5641.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson W R. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]