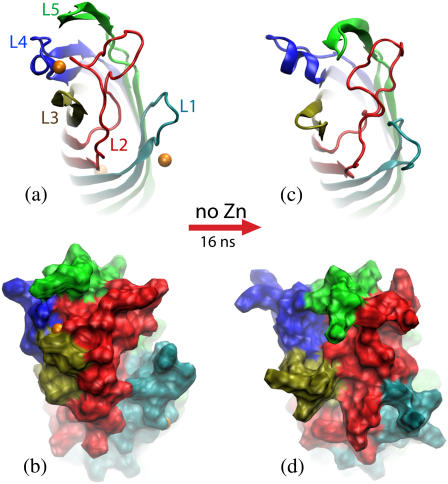
FIGURE 3.
Transformation of the in surfo structure of OpcA simulated in a lipid bilayer membrane. (a and b) The conformation of OpcA in the in surfo x-ray structure. L1 and L2 create a crevice that was proposed to harbor a binding site for proteoglycans (21). Zinc ions (orange), resolved in the x-ray structure, are shown as van der Waals spheres. (c and d) The conformation of the in surfo model after 16-ns equilibration in a lipid bilayer carried out in the absence of the zinc ions. In this simulation, the crevice between L1 and L2 disappeared after ∼8 ns. The protein structure is shown in cartoon (a and c) and molecular surface (b and d) representations. The loops are shown in different colors.