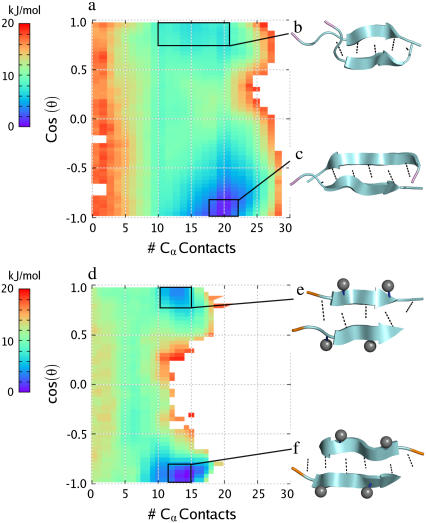
FIGURE 3.
Two-dimensional PMF plots at T = 300 K for the two homodimers studied: (a) the Aβ16–22 peptide and (d) the Aβ16–20m inhibitor. In each plot, the free energy is plotted in units of kJ/mol versus number of interpeptide Cα contacts (x axis) and the cosine of θ, the angle between the two peptides, defined as the angle between the vectors connecting Cα atoms of residues 17 and 19 in the Aβ16–20m inhibitor and residues 18–20 in the Aβ16–22 peptide. The boxes denote the region of phase space representing the lowest energy basins in which conformational clustering was performed. (b–c, e–f) The resulting representative structures from the most populated clusters within these basins are shown on the right. Methyl groups are shown as gray spheres, backbone interpeptide hydrogen bonds are shown as dotted lines, and the N-terminus of each strand is highlighted in orange for the Aβ16–20m inhibitor and in violet for the Aβ16–22 peptide.