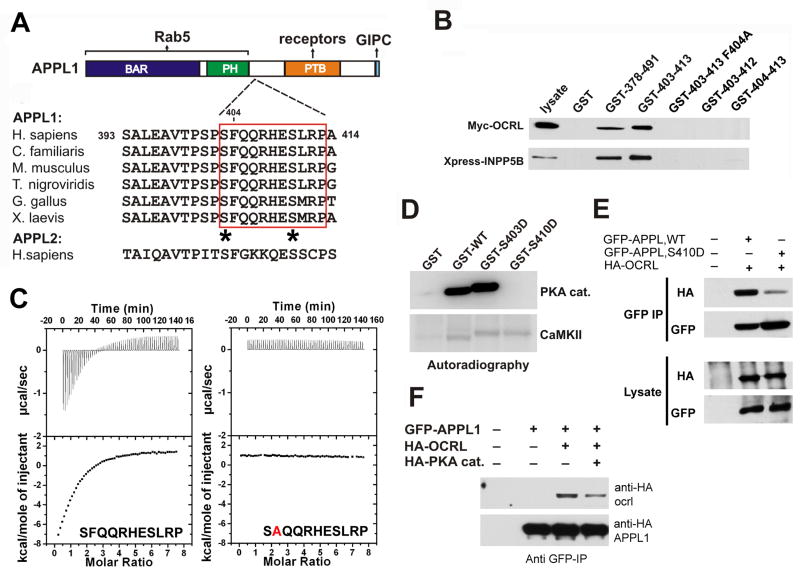
Figure 4. Mapping of the binding interfaces between APPL1 and OCRL.
A: Modular structure and interactions of APPL1. The figure also shows an alignment of the 11-mer peptide of human APPL1 with the corresponding sequence of APPL from other species, demonstrating its evolutionary conservation. The sequence is not conserved in human APPL2. Potential phosphorylation sites within the 11-mer peptide are indicated by asterisks. B: GST pulldowns from extracts of Cos-7 expressing either Myc-OCRL or Xpress-INPP5B define an 11 amino acid (residues 403–413) stretch of human APPL1 as the minimal region necessary and sufficient for binding. Bound proteins were identified by western blotting. Numbers indicate the amino acid boundaries of the APPL1 fragment fused to GST. The F404A mutation abolished binding. C: ITC analysis of the binding of the wildtype APPL1 11-mer peptide and of the F404A-mutant peptide. Raw data are shown in the upper panels, and plots of the total heat released as a function of the molar ratio of each ligand are shown in the bottom panels. D: GST-fusions of the wildtype or mutant (S403D and S410D) minimal binding region (11-mer peptide) were incubated with the catalytic subunit of PKA or with type II Ca2+-calmodulin dependent kinase (CamKII) in the presence of γ-32P-ATP. Proteins were separated by SDS-PAGE and the corresponding autoradiography is shown. E: Co-immunoprecipitation of full length wildtype and mutant (S410D) APPL1 with OCRL. F: PKA overexpression impairs binding of APPL1 to OCRL in vivo. For E and F Cos-7 cells were transfected with the indicated expression constructs. Protein complexes were immunoprecipitated using anti-GFP antibody and the starting lysates and the immunoprecipitates were analyzed by western blotting.