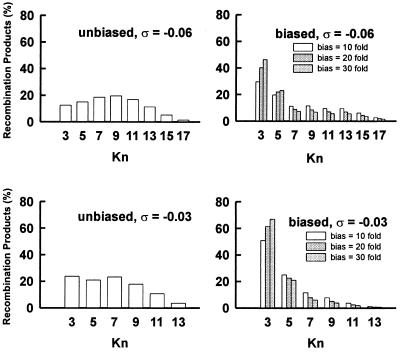
Figure 6.
Calculated knot distributions based on the biased slithering model for intramolecular site-specific recombination of a supercoiled Int substrate. Histograms on the left show the distribution of knots obtained for cases in which there are no sequence-dependent preferences for looped regions; profiles on the right correspond to different end-loop biases, ξ, equal to 10, 20, or 30. Model parameters were based on a 4.60-kb DNA substrate divided into 950-bp and 3.65-kb domains. Recombination substrates were considered to be a heterogeneous population of superhelical molecules with mean values of the linking difference corresponding to high (σ = −0.06) or low (σ = −0.03) superhelix density; this heterogeneity was assumed to obey a Gaussian distribution with variances equal to the experimentally measured values obtained from topoisomer distributions of plasmid substrates. Conformations in which a superhelix end loop was partially occupied by the biased DNA segment were weighted according to their elastic energy, as given in Eq. 1.