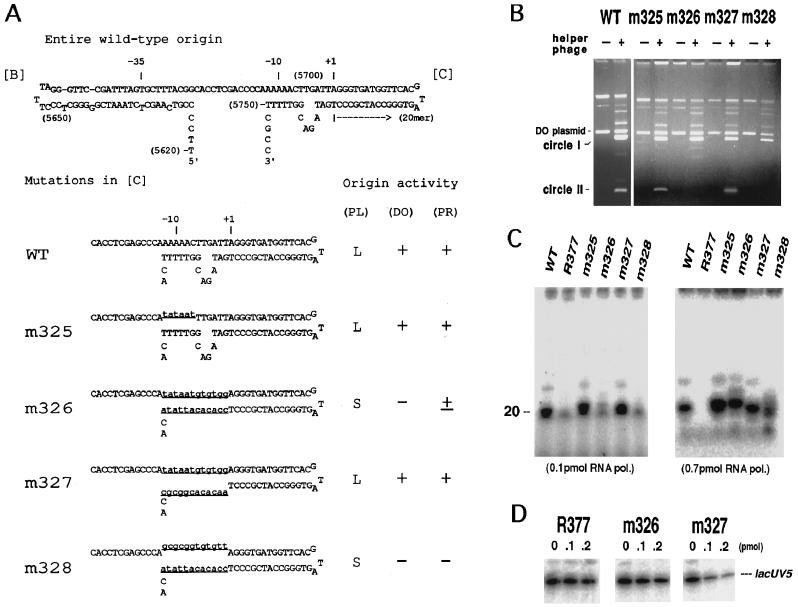
Figure 3.
Origin activity of base-substitution mutants in the −10 region of the minus-strand origin. (A) A possible secondary structure of the wild-type and mutant origins. The curved arrow represents the primer RNA. Numerals in parentheses are f1 nucleotide numbers (11). The altered nucleotides in mutants are shown by underlined lowercase type. Activity of the origins, determined by three methods, plaque size of the phage carrying the mutation, double-origin plasmid assay (3), and primer synthesis in vitro, is qualitatively indicated on the right. (PL), plaque size; L, large; S, small. The results of double-origin plasmid assay (DO) and primer RNA synthesis (PR) are shown in B and C, respectively. For explanation of double-origin plasmid assay, see the text. (B) Results of double-origin plasmid assay for the wild-type, m325, m326, m327, and m328 minus-strand origin. +, Infected with f1; −, uninfected. The positions of solid circles of the parental plasmid (DO plasmid) and its two derivatives, circle I and circle II, are indicated on the left. Intensity of the circle II band is a measure of activity of the mutant origin. (C) In vitro synthesis of primer RNA on mutant origins. Viral ssDNA (0.1 pmol) extracted from mutant phages was used as templates as indicated. RNA synthesized with 0.1 pmol (Left) or 0.7 pmol (Right) of RNA polymerase holoenzyme in a standard reaction was electrophoresed in 12% (Left) or 15% (Right) polyacrylamide gel containing 8 M urea and radioautographed. (D) In vitro mixed RNA synthesis using lacUV5 promoter and mutant origins. The reaction contained 0.1 pmol of lacUV5–3, a DNA fragment carrying lacUV5 promoter, and 0–0.2 pmol of competitor (viral ssDNA) as indicated.