Abstract
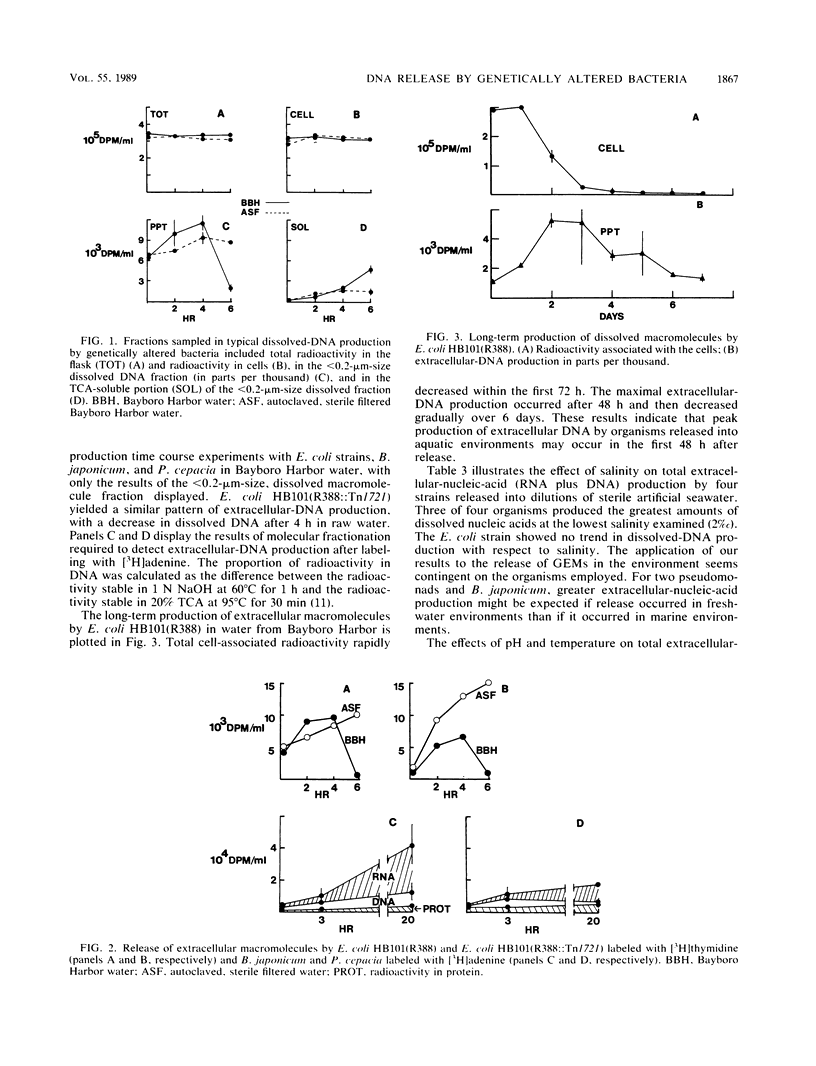
The factors which affect the production of extracellular DNA by genetically altered strains of Escherichia coli, Pseudomonas aeruginosa, Pseudomonas cepacia, and Bradyrhizobium japonicum in aquatic environments were investigated. Cellular nucleic acids were labeled in vivo by incubation with [3H]thymidine or [3H]adenine, and production of extracellular DNA in marine waters, artificial seawater, or minimal salts media was determined by detecting radiolabeled macromolecules in incubation filtrates. The presence or absence of the ambient microbial community had little effect on the production of extracellular DNA. Three of four organisms produced the greatest amounts of extracellular nucleic acids when incubated in low-salinity media (2% artificial seawater) rather than high-salinity media (10 to 50% artificial seawater). The greatest production of extracellular nucleic acids by P. cepacia occurred at pH 7 and 37 degrees C, suggesting that extracellular-DNA production may be a normal physiologic function of the cell. Incubation of labeled P. cepacia cells in water from Bimini Harbor, Bahamas, resulted in labeling of macromolecules of the ambient microbial population. Collectively these results indicate that (i) extracellular-DNA production by genetically altered bacteria released into aquatic environments is more strongly influenced by physiochemical factors than biotic factors, (ii) extracellular-DNA production rates are usually greater for organisms released in freshwater than marine environments, and (iii) ambient microbial populations can readily utilize materials released by these organisms.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borenstein S., Ephrati-Elizur E. Spontaneous release of DNA in sequential genetic order by Bacillus subtilis. J Mol Biol. 1969 Oct 14;45(1):137–152. doi: 10.1016/0022-2836(69)90216-2. [DOI] [PubMed] [Google Scholar]
- Deflaun M. F., Paul J. H., Davis D. Simplified method for dissolved DNA determination in aquatic environments. Appl Environ Microbiol. 1986 Oct;52(4):654–659. doi: 10.1128/aem.52.4.654-659.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman J. A., McManus G. B. Do bacteria-sized marine eukaryotes consume significant bacterial production? Science. 1984 Jun 15;224(4654):1257–1260. doi: 10.1126/science.224.4654.1257. [DOI] [PubMed] [Google Scholar]
- Morrison W. D., Miller R. V., Sayler G. S. Frequency of F116-mediated transduction of Pseudomonas aeruginosa in a freshwater environment. Appl Environ Microbiol. 1978 Nov;36(5):724–730. doi: 10.1128/aem.36.5.724-730.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Jeffrey W. H., David A. W., Deflaun M. F., Cazares L. H. Turnover of extracellular DNA in eutrophic and oligotrophic freshwater environments of southwest Florida. Appl Environ Microbiol. 1989 Jul;55(7):1823–1828. doi: 10.1128/aem.55.7.1823-1828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Jeffrey W. H., DeFlaun M. F. Dynamics of extracellular DNA in the marine environment. Appl Environ Microbiol. 1987 Jan;53(1):170–179. doi: 10.1128/aem.53.1.170-179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H. Use of hoechst dyes 33258 and 33342 for enumeration of attached and planktonic bacteria. Appl Environ Microbiol. 1982 Apr;43(4):939–944. doi: 10.1128/aem.43.4.939-944.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann B., Søndergaard M. Measurements of diel rates of bacterial secondary production in aquatic environments. Appl Environ Microbiol. 1984 Apr;47(4):632–638. doi: 10.1128/aem.47.4.632-638.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saye D. J., Ogunseitan O., Sayler G. S., Miller R. V. Potential for transduction of plasmids in a natural freshwater environment: effect of plasmid donor concentration and a natural microbial community on transduction in Pseudomonas aeruginosa. Appl Environ Microbiol. 1987 May;53(5):987–995. doi: 10.1128/aem.53.5.987-995.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt R., Bernhard E., Mattes R. Characterisation of Tn1721, a new transposon containing tetracycline resistance genes capable of amplification. Mol Gen Genet. 1979 Apr 17;172(1):53–65. doi: 10.1007/BF00276215. [DOI] [PubMed] [Google Scholar]
- Ward J. M., Grinsted J. Mapping of functions in the R-plasmid R388 by examination of deletion mutants generated in vitro. Gene. 1978 Apr;3(2):87–95. doi: 10.1016/0378-1119(78)90053-7. [DOI] [PubMed] [Google Scholar]



