Abstract
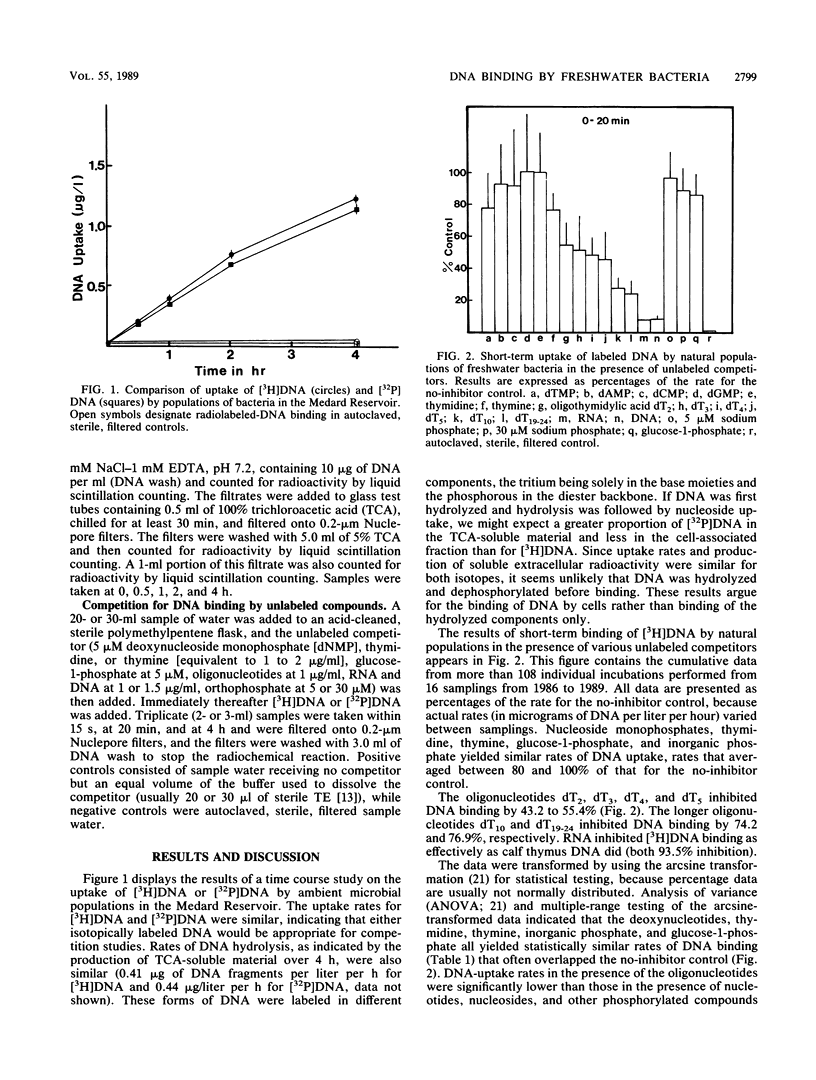
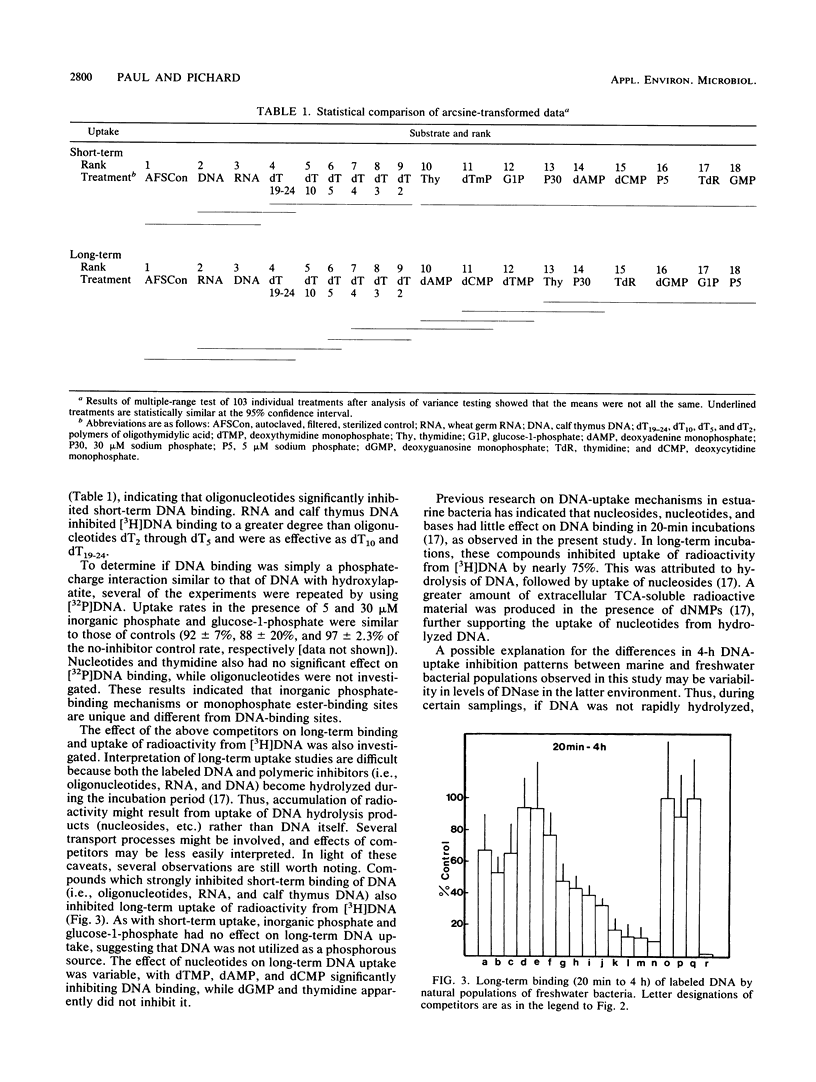
The substrate specificity of the DNA-binding mechanism(s) of bacteria in a Florida reservoir was investigated in short- and long-term uptake studies with radiolabeled DNA and unlabeled competitors. Thymine oligonucleotides ranging in size from 2 base pairs to 19 to 24 base pairs inhibited DNA binding in 20-min incubations by 43 to 77%. Deoxynucleoside monophosphates, thymidine, and thymine had little effect on short-term DNA binding, although several of these compounds inhibited the uptake of the radiolabel from DNA in 4-h incubations. Inorganic phosphate and glucose-1-phosphate inhibited neither short- nor long-term binding of [3H]- or [32P]DNA, indicating that DNA was not utilized as a phosphorous source in this reservoir. RNA inhibited both short- and long-term radiolabeled DNA uptake as effectively as unlabeled DNA. Collectively these results indicate that aquatic bacteria possess a generalized nucleic acid uptake/binding mechanism specific for compounds containing phosphodiester bonds and capable of recognizing oligonucleotides as short as dinucleotides. This binding site is distinct from nucleoside-, nucleotide-, phosphomonoester-, and inorganic phosphate-binding sites. Such a nucleic acid-binding mechanism may have evolved for the utilization of extracellular DNA (and perhaps RNA), which is abundant in many marine and freshwater environments.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Kahn M. E., Barany F., Smith H. O. Transformasomes: specialized membranous structures that protect DNA during Haemophilus transformation. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6927–6931. doi: 10.1073/pnas.80.22.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noteborn M., Venema G., Kooistra J. Effect of ethylenediaminetetraacetic acid on deoxyribonucleic acid entry and recombination in transformation of a wild-type strain and a rec-1 mutant of Haemophilus influenzae. J Bacteriol. 1981 Mar;145(3):1189–1195. doi: 10.1128/jb.145.3.1189-1195.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Deflaun M. F., Jeffrey W. H. Mechanisms of DNA utilization by estuarine microbial populations. Appl Environ Microbiol. 1988 Jul;54(7):1682–1688. doi: 10.1128/aem.54.7.1682-1688.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Jeffrey W. H., DeFlaun M. F. Dynamics of extracellular DNA in the marine environment. Appl Environ Microbiol. 1987 Jan;53(1):170–179. doi: 10.1128/aem.53.1.170-179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Danner D. B., Deich R. A. Genetic transformation. Annu Rev Biochem. 1981;50:41–68. doi: 10.1146/annurev.bi.50.070181.000353. [DOI] [PubMed] [Google Scholar]
- Stewart G. J., Carlson C. A. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211–235. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]


