Abstract
A protein-protein Förster resonance energy transfer (FRET) system, employing probes at multiple positions, was designed to specifically monitor the dissociation of the H2A-H2B dimer from the nucleosome core particle (NCP). Tryptophan donors and Cys-AEDANS acceptors were chosen because, in comparison to fluorophores used in previous NCP FRET studies, they: 1) are smaller and less hydrophobic which should minimize perturbations of histone and NCP structure; and 2) have an R0 of 20 Å, which is much less than the dimensions of the NCP (~50 Å width and ~100 Å diameter). CD and FL equilibrium protein unfolding titrations indicate that the donor and acceptor moieties have minimal effects on the stability of the H2A-H2B dimer and (H3-H4)2 tetramer. NCPs containing the various FRET pairs were reconstituted with the 601 artificial positioning DNA sequence. Equilibrium NaCl-induced dissociation of the modified NCPs showed that the 601 sequence stabilized the NCP to dimer dissociation as compared to previous studies using weaker positioning sequences. This finding implies a significant role for the H2A-H2B dimers in determining the DNA sequence dependence of NCP stability. The free energy of dissociation determined from reversible and well-defined sigmoidal transitions revealed two distinct phases reflecting the dissociation of each H2A-H2B dimer, confirming cooperativity in dimer dissociation. While cooperativity in the association/dissociation of the H2A-H2B dimers has been suggested previously, these data allow its quantitative description. The protein-protein FRET system was then used to study the effects of the histone variant H2A.Z on NCP stability; previous studies have reported both destabilizing and stabilizing effects. Comparison of the H2A and H2A.Z FRET NCP dissociation transitions suggest a slight increase in stability but a significant increase in cooperativity for dimer dissociation from H2A.Z NCPs. Thus, the utility of this protein-protein FRET system to monitor the effects of histone variants on NCP dynamics has been demonstrated, and the system appears equally well-suited for dissection of the kinetic processes of dimer association and dissociation from the NCP.
Keywords: FRET, fluorescence, thermodynamics, histone variants, chromatin
The nucleosome core particle (NCP) is the fundamental repeating unit for packaging DNA into chromatin. Approximately 150 bp of DNA are wrapped around an octamer of histones, containing two copies each of H2A, H2B, H3 and H4. Although it was initially thought to be a static, highly regular packaging system, the nucleosome is now known to be a dynamic structure.1–3 Changes in the properties of the NCP are critical in the regulation of DNA-dependent processes, such as transcription, replication, recombination and repair. The misregulation of NCP structure and function leads to a range of diseases, particularly cancer.4–6
The histones of the core nucleosome contain N-terminal tails and a conserved three helix heterodimerization motif, as well as C-terminal extensions in the H2A-H2B dimer.7 The H3-H4 dimer further oligomerizes to a tetramer through a four helix bundle comprised of the C-terminal helical regions of H3 and H3′. In the NCP, the C-terminal regions of H2B and H4 form two additional four helix bundles within the octamer.8; 9 Salt bridges and hydrogen bonds between buried charged side chains stabilize the four-helix bundles, as well as burial of hydrophobic surface area. The L1 loop and C-terminus of H2A also participate in important intermolecular interfaces. The L1 loops of H2A and H2A′ interact with each other through multiple hydrogen bonds; it has been hypothesized that stabilization arising from these loop interactions may contribute to the cooperative association of the H2A-H2B dimers in formation of the NCP.8; 10 The “docking domain” of the C-terminal tail of H2A also makes extensive contacts with the H3-H4 dimer and contributes to the helical ramp for the DNA.
The process of NCP assembly from purified proteins is similar to that observed in the cell.11; 12 The (H3-H4)2 tetramer binds first and positions the central portion of the DNA wrapped in the NCP. The H2A-H2B dimers bind to surfaces on each side of the tetramer, completing the two helical ramps that position the 1.65 superhelical turns of DNA. In the cell, histone deposition is controlled by histone chaperones such as CAF-1, RCAF and NAP-1 (for review13). Cell-free reconstitutions of the NCP generally rely on the different sensitivities to ionic strength of the assembly/disassembly steps. The NaCl-induced disassembly process is shown in Scheme 1: At NaCl concentrations approaching physiological ionic strength, the mobility of the DNA ends increase. As the NaCl concentrations are increased, the H2A-H2B dimers dissociate, and finally the (H3-H4)2 tetramer. Dissociation of the H2A-H2B dimers begins at ~0.4 M NaCl for many DNA sequences, including the commonly used sea urchin 5S gene sequence. The (H3-H4)2 tetramer dissociates above 1.4 M NaCl.11; 14; 15 The transient formation of a hexameric intermediate, shown in brackets in Scheme 2, is not directly detected by most current methods.
Scheme 2.
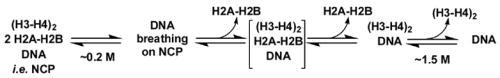
Salt-induced NCP unfolding
The intrinsic dynamics of the NCP result from movement of either the DNA or the histone oligomers in the absence of extrinsic proteins. Widom and colleagues showed that target sites wrapped inside the NCP are transiently exposed to DNA-binding proteins of various sizes.16; 17 Equilibrium and kinetic studies of DNA-DNA FRET (Förster resonance energy transfer) systems have been used to monitor the dynamic mobility of the DNA in the NCP.18–20 Spontaneous wrapping and unwrapping of the DNA ends as well as larger scale movements near the dyad axis occur on timescales from tens to hundreds of milliseconds. The NCP is in a closed state ~95% of the time, but unwrapping permits access of non-histone proteins to various DNA sequences. Clearly, the nucleosome is in a constant flux between wrapped and unwrapped states. The magnitude and rate of this dynamic flux is influenced by extrinsic factors, including the incorporation of histone variants, post-translational modifications, histone chaperones and ATP-dependent remodeling complexes. FRET experiments have been and will continue to be a powerful tool to dissect NCP dynamics.
Histone variants constitute a small, but essential fraction of a cell’s histone complement and play distinct functional, and presumably structural, roles in chromatin dynamics.21–25 The greatest number of variants have been identified for H2A including the major H2A (H2A.1), H2A.Z, H2A.X, H2A-Bbd, and MacroH2A. The variants constitute ≤ 20% of the total H2A population and vary from 60% to 90% sequence identity with the major H2A. The main differences relative to H2A.1 are the sequence and length of the C-terminal α-helix and extended tail as well as significant changes in the L1 loop. Of these variants, H2A.Z has the highest abundance in the proteome and has been highly studied. While H2A.Z deficient strains of Saccharomyces cerevisiae are viable, this variant is essential in all higher eukaryotes.23 Various studies have implicated H2A.Z in both transcriptional activation and repression (e.g. 26–31). An emerging view is that H2A.Z plays a role in defining and maintaining the boundaries of heterochromatin.32; 33
The X-ray crystal structure of H2A.Z NCP is very similar to that of NCPs containing the major H2A with minimal changes in protein-DNA contacts.10 Two regions exhibited subtle structural differences that may influence the stability and dynamics of individual NCPs: 1) The interface between the C-terminal docking domain of H2A which binds the H3-H4 dimer; and 2) The H2A-H2A′ L1 loop interactions. Firstly, the sequence differences in the C-terminal docking domains caused small localized changes, such that three hydrogen bonds observed in the H2A NCP were absent in the H2A.Z NCP structure—which may potentially destabilize the H2A.Z NCP. Secondly, the relative orientations of the L1 and L1′ loops are altered, changing the intermolecular hydrogen bonding between H2A-H2A′ and H2A.Z-H2A.Z′—two and six hydrogen bonds, respectively. These changes may increase the binding cooperativity for H2A.Z-H2B dimers and disfavor “mixed” NCPs containing both major H2A and H2A.Z histones.10; 15
There is some controversy about the relative stability of H2A and H2A.Z NCPs. Initial studies suggested that H2A.Z decreased the stability of the NCP to salt induced dissociation.34 Subsequent studies, using salt-induced dissociation,15 and sucrose gradient or hydroxyapatite chromatin fractionations35 have suggested minor stabilization upon incorporation of unacetylated H2A.Z while temperature-dependent NCP mobility suggests that H2A.Z is destabilizing.36 Additional biophysical studies are required to clarify the determinants of the relative stabilities of NCPs containing H2A and H2A.Z.
To further characterize NCP assembly/disassembly with major and variant histones, a FRET system was developed to monitor protein-protein interactions within the NCP. Unique Trp donors were engineered into the H3 and H4 histones. Unique Cys residues, targets for IAEDANS modification, were engineered into H2A and H2B. Five different FRET Trp donor/Cys-AEDANS acceptor pairs were reconstituted into NCPs with the 601 artificial positioning sequence.37 NCP assembly/disassembly was monitored by salt-induced dissociation. Two important features of this FRET system permitted an estimation of the free energy for H2A-H2B dissociation. Firstly, FRET is sensitive to the dissociation of the dimer and relatively insensitive to other transitions such as DNA breathing or tetramer dissociation. Secondly, the salt-induced dissociation of the H2A-H2B dimer is highly reversible.
RESULTS
Design of components of the NCP FRET system
The histone octamer wraps ~150 bp of DNA, and many sequences can populate multiple translational and rotational positions. To ensure a homogeneous NCP population, the strong positioning 601 sequence identified by the Widom lab was used.37; 38 The affinity of the 601 sequence for the histone octamer is ~150 times greater than that of the well-characterized positioning sequence from the sea urchin 5S rRNA gene, used in a previous NCP FRET equilibrium study.15
Previous NCP FRET systems have used derivatives of coumarin, fluorescein, Cy3 and Cy5 with Förster distances of 50 to 60 Å.15; 18–20; 39; 40 The Trp to Cys-AEDANS pair was chosen because: 1) the relatively small size and hydrophobic surface area of the fluorophores will minimize any tendency of the fluorophores to interact with hydrophobic patches on or near the surface of the proteins; and 2) the R0 of 20 Å41 is substantially less than the dimensions of the NCP (~50 Å width and ~100 Å diameter).
The sites for the FRET donors (Trp on H3 and H4) and acceptors (Cys-AEDANS on H2A and H2B), depicted in Table 1 and Figure 1, were chosen based on inspection of the NCP crystal structure.8 The choices were based on four factors: 1) the absence of Trp residues in all four histones and Cys in H2A, H2B and H4; 2) high solvent accessibility of the WT side chain to permit rotational freedom of the fluorophore; 3) hydrophobicity of the WT residue to minimize structural perturbation; 4) proximity of the Cβ atoms of the donor and acceptor sites. The H2A-L108C mutation is at the beginning of the C-terminal tail, which is in close proximity to the structured regions of the histone octamer. The H2B-S109C mutation is on the surface of the C-terminal helix, beyond the canonical histone fold. The H3-F78W mutation is the C-terminal residue of the α1 helix of the canonical histone fold. Because of distance considerations, Trp residues were introduced on the surface of the central α2 helix of the H4 histone fold by mutation of Leu-49 and Val-60. Five protein-protein FRET NCPs (Table 1) were designed to monitor the dissociation of H2A-H2B dimer. The distances from one donor to each acceptor (D-A and D-A′) are similar for FRET NCPs with H2A Cys-AEDANS acceptors, but dissimilar for H2B-acceptor NCPs.
Table 1.
Distances between the Cβ atoms of the residues mutated for introduction of the Trp donors and Cys-AEDANS acceptors.a
| Distances from H3-H4 to: | |||
|---|---|---|---|
| FRET Pair | proximal H2A-H2B | distal H2A-H2B | Average distance and % FRET Efficiencyb |
| H3-78W to H2A-108Cys-AEDANS | 32.9 & 33.0 | 35.5 | 34.2 Å, 3.8% |
| H4-49W to H2A-108Cys-AEDANS | 30.2 & 30.5 | 28.4 | 29.4 Å, 8.8% |
| H4-60W to H2A-108Cys-AEDANS | 23.8 & 24.7 | 26.6 & 26.9 | 25.5 Å, 18.9% |
|
| |||
| H3-78W to H2B-109Cys-AEDANS | 19.6 & 20.0 | 48.9 & 49.6 | 51.9 & 0.5% |
| H4-60W to H2B-109Cys-AEDANS | 26.9 & 26.8 | 45.4 & 45.7 | 14.2 & 0.7% |
Distances determined from the coordinates of the NCP X-ray crystal structure, 1AOI.pdb.8 When two distances are shown, this reflects slight differences between the two copies of the histones in the NCP. The proximal H2A-H2B dimer is defined as the one on the same face of the NCP as the H3-H4 Trp donor.
The expected FRET efficiency was calculated from the Förster distance of ~20 Å for the Trp:Cys-AEDANS FRET pair41 and the estimated distances between the Cβ atoms. For H2A-acceptors, the efficiency is calculated for the average distance of the four D-A pairs. For H2B-acceptors, the D-A and D-A′ distances are very different and expected FRET efficiencies are shown for both distances.
Figure 1.
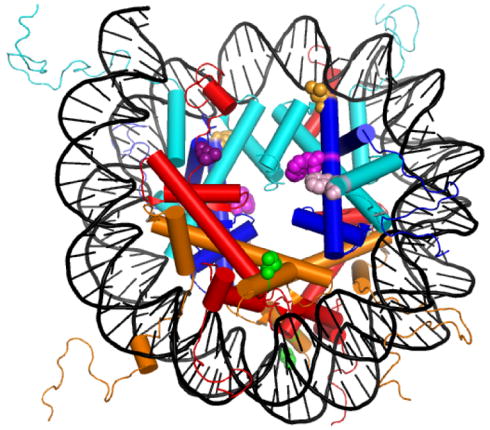
The structure of the NCP (1kx5.pdb)79 showing the sites mutated to introduce FRET fluorophores (Table 1). Histones are colored: H2A, red; H2B, orange; H3, cyan; and H4, blue. Mutations: H2A Leu-108 (dark purple); H2B Ser-109 (green); H3 Phe-78 (pink); H4 Leu-49 (orange), and H4 Val-60 (light purple). This figure was rendered with Pymol (Delano Scientific, LLC, San Carlos, CA).
Fluorescence spectra and anisotropy of the Cys-AEDANS and Trp residues
Fluorescence emission spectra of folded and denaturant-unfolded oligomers were compared to assess the solvent accessibility of the FRET donors and acceptors. The emission wavelength maximum should red shift very little upon the addition of denaturants if the fluorophore is highly exposed to solvent in the native oligomer. The folded H2A-H2B dimers containing the Cys-AEDANS acceptor had emission maxima of 485 to 490 nm; upon unfolding, there was a minimal red shift of 5 nm. The Trp FL of the folded H3-H4 tetramers exhibited emission maxima of 345 to 350 nm, with 5 nm red shifts upon GdmCl-induced unfolding. These data demonstrate that the FRET donors and acceptors have relatively high solvent accessibility in the folded histone oligomers.
If the fluorophores are not restricted by partial burial or interactions with hydrophobic surfaces on the histones or the NCP, they should exhibit a high degree of rotational freedom, which is important for FRET efficiency. The FL anisotropy parameter, r, is a measure of a fluorophore’s rotational freedom. The relative magnitude of the r value depends on the environment of the fluorophore and the size of the complex to which it is attached; i.e. whether it is: 1) buried and rotation is limited by the tumbling of the macromolecular complex; 2) surface exposed and tumbles somewhat faster than the macromolecular complex; or 3) freely rotating in solvent with a rate that is independent of the tumbling time of the complex. Typical r values for immobilized Trp (propylene glycol, −50 °C) are ~0.25.42
The anisotropy of the FRET fluorophores (Table 2) were determined for histones under native and unfolded conditions; the latter should reflect the maximal rotational freedom. The r values for the Cys-AEDANS fluorophores are ~0.06 in the native H2A-H2B dimers and decrease to ~0.02 upon unfolding. Consistent with a larger molecular weight oligomer and a shorter linker length between the fluorophore and the protein, the Trp residues in the H3-H4 tetramer exhibit slightly higher r values: 0.08 to 0.1 and 0.06 to 0.04 in the native and unfolded species, respectively. In sum, the fluorescence anisotropies and emission maxima are consistent with FRET fluorophores that are solvent-exposed with substantial rotational freedom.
Table 2.
Parameters describing the equilibrium stabilitya and fluorescence anisotropyb of the histones containing FRET donors and acceptors.
| Histone Oligomer | ΔG°(H2O) (kcal mol−1) | m value (kcal mol−1M−1) | Cm | Anisotropy (r) | ||
|---|---|---|---|---|---|---|
| F | U | NCP | ||||
|
H2A-H2B dimerc
|
||||||
| WT | 11.8 | 2.9 | 1.7 | -- | -- | -- |
| H2A-108Cys-AEDANS | 12.1 (0.3) | 3.1 (0.1) | 1.7 | 0.06 | 0.02 | 0.04 |
| H2B-109Cys-AEDANS | 11.9 (0.3) | 2.9 (0.2) | 1.7 | 0.06 | 0.02 | 0.05 |
|
| ||||||
|
H3-H4 dimerd
|
||||||
| WT | 11.8 | 2.6 | 1.7 | -- | -- | -- |
| H3-78W | 11.7 (0.5) | 2.9 (0.3) | 1.5 | 0.10 | 0.06 | 0.11 |
| H4-49W | 12.8 (0.5) | 3.1 (0.2) | 1.7 | 0.08 | 0.04 | 0.07 |
| H4-60W | 11.7 (0.2) | 2.5 (0.1) | 1.8 | 0.08 | 0.04 | 0.14 |
The ΔG°(H2O) is the free energy of unfolding in the absence of denaturant; the m values reflect the slope of the unfolding transition (Eq. 1). CM is the transition midpoint, i.e. the denaturant concentration where [U] = 0.5·[Monomer]Total. The values in parentheses are the error at one standard deviation determined from rigorous error analysis of the global fits. Buffer conditions for the histone data in this table: 0.2 M KCl, 0.1 mM EDTA, 20 mM KPi pH 7.2, 25°C. Conditions for NCP anisotropy are given in legend of Figure 5.
The r values for the folded oligomers, F; the unfolded monomers, U; and the NCP at 0 M NaCl.
The urea-induced unfolding parameters for the WT H2A-H2B dimer are from reference.67 The Cm values are for transitions at 10 μM monomer.
The values represent the stability of the H3-H4 dimer to GdmCl denaturation in the presence of 1 M TMAO. The Cm values are for transitions at 4 μM monomer. The parameters for WT H3-H4 dimers are from reference.43
The stabilities of the engineered histone oligomers are not significantly altered
It is important to assess the effect of the FRET fluorophores on histone stability; changes in protein stability may indicate significant structural alterations, which could alter NCP stability and dynamics. The stability to denaturant-induced unfolding of the engineered histone oligomers was determined by monitoring far-UV circular dichroism (CD) and tyrosine or tryptophan FL. Like the WT histones, the equilibrium unfolding of the FRET-engineered histones was highly reversible, two-state and protein concentration dependent (see Supplementary data). The reversibility and absence of hysteresis was indicated by the coincidence of transitions of folded protein titrated with denaturant-unfolded protein and unfolded protein titrated with folded protein. Two-state unfolding was indicated by the similarity of far-UV CD and FL transitions, which report on secondary and tertiary/quaternary structure, respectively. The high quality of the global fits of the protein-concentration dependence of the data provided further support for the absence of equilibrium intermediates.
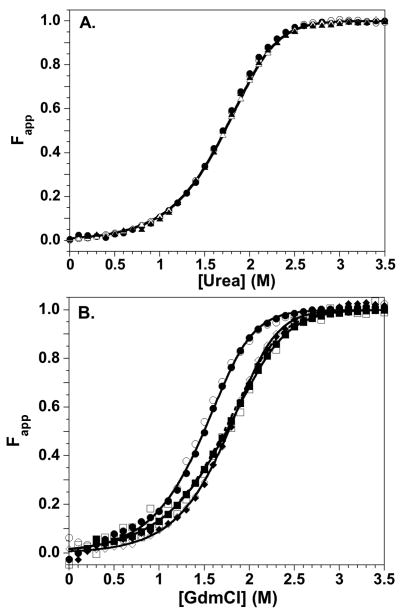
The stabilities of the Cys-AEDANS H2A-H2B variants were determined from urea-induced unfolding titrations at 5 and 10 μM monomer, monitored by both far-UV CD and Tyr FL. The fluorescence of the Cys-AEDANS moiety was not sensitive to unfolding, as expected for a solvent exposed, highly mobile fluorophore. For each variant, a data set of six titrations (two monomer concentrations, two spectroscopic probes with folding and unfolding titrations) was analyzed by global fitting. The fitted parameters are given in Table 2, and representative Fapp transitions are shown in Figure 2A. The stabilities of the modified H2A-H2B dimers are very similar to the WT dimer, exhibiting similar transition midpoints, CM, ΔG°(H2O) and m values. These data indicate that the FRET acceptors cause no significant changes in structure.
Figure 2.
Equilibrium stability of the WT and engineered histones. CD data, open symbols; FL data, closed symbols. Equilibrium titration data were collected from 0 to 4 M denaturant, but the transition region is expanded for clarity. A) Urea-induced denaturation of H2A-H2B dimers at 10 μM monomer. H2A-108Cys-AEDANS/H2B, circles; H2A/H2B-109Cys-AEDANS, triangles. The transition for WT H2A-H2B is represented by the solid line. B) GdmCl-induced denaturation of the H3-H4 oligomers at 4 μM monomer in 1 M TMAO. H3-78W/H4, circles; H3/H4-49W, diamonds; H3/H4-60W, squares. The solid lines represent global fits of the entire data sets for the histone mutants. The WT H3-H4 transition is represented by the dashed line. Conditions: 200 mM KCl, 0.1 mM EDTA, 20 mM KPi pH 7.2, 25°C.
The H3-H4 Trp variants were denatured with guanidinium chloride, GdmCl, as in previous studies on the WT histones.43 The instability of the H3-H4 oligomers prevented the precise definition of native baselines, which is necessary for quantitative analyses; therefore the osmolyte trimethylamine-N-oxide (TMAO) was used. TMAO stabilizes the native state and extends the native, pre-transition baseline region in equilibrium denaturation studies.43–46
Denaturation of the H3-H4 variants at 2 and 4 μM monomer was monitored by far-UV CD and Trp FL. Data sets of 5 to 6 titrations were analyzed by global fits for each H3-H4 Trp variant. Despite their solvent-exposure, the FL of the Trp residues was sensitive to the unfolding transition. As suggested by FL anisotropy, the Trp residues may be more closely tethered to the central histone fold than the Cys-AEDANS residues on H2A and H2B. The far-UV CD signal of WT H3-H4 was insensitive to the dissociation of the tetramer to native heterodimers, and the unfolding transitions were well described by a two-state dimeric mechanism.43 The FL from multiple Tyr residues in WT H3-H4 reported on both tetramer dissociation and dimer unfolding, requiring fits to a three-state mechanism. The FL of the single Trp residues are insensitive to tetramer dissociation, which is consistent with their lack of proximity to the H3-H3′ tetramer interface. The transitions monitored by Trp FL and far-UV CD were coincident for the variant H3-H4 oligomers, and their protein concentration dependence was well-described by global fits to a two-state dimer unfolding model (Supplementary Material Figure 2). Fits to a three-state model with tetramer dissociation and dimer unfolding did not significantly improve the reduced Chi-squared values. The fitted parameters for the two-state global fits and representative Fapp transitions are presented in Table 2 and Figure 2B, respectively. The ΔG°(H2O) values for the H3-78W and H4-60W heterodimers were identical to that of the WT H3-H4 dimer; the m values are slightly increased, resulting in minor changes in the CM values. Trp-49 in the α2 helix of H4 slightly stabilizes the heterodimer as assessed by the ΔG°(H2O) and m values, although the CM value is identical to WT H3-H4. In summary, introduction of Trp residues into H3 and H4 has minimal effects on the stability, and presumably the structure, of the H3-H4 heterodimer.
Salt induced dissociation of the WT NCP
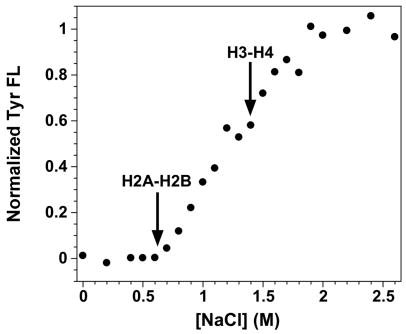
As in protein folding studies, perturbation methods are used to characterize NCP stability; the most commonly employed perturbant is salt, specifically NaCl. The stabilities of unmodified NCPs have been monitored previously by the intrinsic FL of the 30 Tyr residues distributed throughout the eight core histones.14; 15; 47 At low ionic strength, Tyr FL is quenched by the DNA in the fully assembled NCP. Salt-induced dissociation of the histone oligomers results in an increase in the Tyr FL intensity. A minor transition is observed around 0.2 M NaCl from increased DNA breathing (Scheme 1). Two major transitions occur at higher NaCl concentrations and have been shown to arise from dissociation of the H2A-H2B dimers and then the (H3-H4)2 tetramer.
The 601 sequence has been used for kinetic FRET studies,18; 19 but the salt-dependent dissociation of 601-NCPs with unmodified histones hasn’t been reported. For comparison to published studies with other DNA sequences, the salt-induced disassembly of NCPs reconstituted from WT histones and the 601 sequence were monitored by Tyr FL (Fig. 3). Multiple transitions were observed as described previously (for example, 14; 47). The low salt transition known to be DNA breathing (Scheme 1) is not apparent. The first major transition, which previous studies have demonstrated to be H2A-H2B release, begins at ~0.6 M NaCl, compared to 0.3 to 0.4 M for NCPs reconstituted with the commonly used sea urchin 5S gene sequence.15 This suggests that the H2A-H2B dimer forms a more stable protein-DNA complex in the 601 NCPs. A recent DNA-DNA NCP FRET system also suggested the potential for different DNA sequences to alter the affinity of the H2A-H2B dimers for the (H3-H4)2·DNA complex.39 The second Tyr FL transition, corresponding to (H3-H4)2 dissociation, begins at ~1.5 M NaCl. Complete histone dissociation is achieved by 2 M NaCl for NCPs with either 601 or 5S rRNA DNA sequences.
Figure 3.
NaCl-induced dissociation of NCPs reconstituted with the 601 DNA sequence and recombinant WT histones monitored by Tyr FL. Arrows indicate the beginning of the transitions corresponding to the dissociation the H2A-H2B dimers and H3-H4 tetramer. Conditions: 250 nM NCP, 0.1 mM EDTA, 0.1 mM β-ME, 20 mM Tris-Cl pH 7.6, 25°C.
Characterization of FRET nucleosome core particles
NCPs were reconstituted with the appropriate FRET donors and acceptors (Table 1) as well as with only donor- or acceptor-containing histones. Native polyacrylamide gel electrophoresis was used to assess the homogeneity of the NCP reconstitutions. PAGE mobility is also a sensitive probe of the integrity of the NCP.48 DNA was visualized by ethidium bromide staining (Figure 4A). Coomassie Blue staining showed that the protein component co-migrated with the DNA (data not shown). Similar gel mobility and homogeneity were observed for unmodified NCPs and those containing only donors or acceptors (data not shown).
Figure 4.
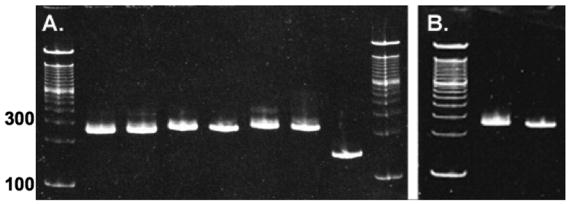
NCPs reconstituted with 149 bp 601 DNA electrophoresed on 5% native acrylamide gels and stained with EtBr. A. NCPs with major H2A. From left to right: 100 bp ladder; H4-60W/H2A-108C-AEDANS; H4-60W/H2B-109C-AEDANS; H4-49W/H2A-108C-AEDANS; H4-49W/H2B-109C-AEDANS; H3-78W/H2A-108C-AEDANS; H3-78W/H2B-109-C-AEDANS; 149 bp 601 DNA. B. NCPs with H2A.Z. From left to right: 100 bp ladder; H3-78W/H2B-109C-AEDANS; H4-60W/H2B-109C-AEDANS.
The salt-dependent fluorescence of NCPs containing only donor or acceptor fluorophores was examined. There was no significant FL change for any of the Trp donors between 0 and 2.4 M NaCl. This demonstrates that the FL of the Trp donors is insensitive to tetramer dissociation, which is consistent with their placement at distances sufficiently distant from the DNA to avoid the quenching observed for intrinsic Tyr residues (Figure 3). This insensitivity is a requirement for a FRET donor, allowing donor FL changes to be attributed solely to quenching by proximity to the acceptor, and not to local conformational changes.
The acceptor-only NCPs showed minor salt dependent FL changes. H2A-108Cys-AEDANS NCPs exhibited a small linear FL increase of ~10% between 0 and 2 M NaCl. This slight salt dependence most likely reflects a solvent effect, rather than any structural transitions. The FL of the H2B-109Cys-AEDANS decreased ~20% between 0.5 and 1.5 M NaCl, suggesting a minor FL response to histone dissociation or a conformational change. However, this FL change is much less than that observed in the FRET experiments (see below). Quantitative analyses of the salt-induced dissociation of FRET NCPs with the H2B acceptor focused on Trp donor data to avoid complications from this small acceptor FL change.
The anisotropy of NCPs containing only donor or acceptor fluorophores are given in Table 2. In the NCP, H2A and H2B Cys-AEDANS residues exhibit r values that are intermediate between those of the folded dimer and unfolded monomer. The Trp r values in the NCP decrease slightly (H4-49W) or exhibit slight to moderate increases (H4-60W and H3-78W, respectively) relative to the folded (H3-H4)2 tetramer.
In summary, native gel electrophoresis and fluorescence anisotropy demonstrate that the FRET probes do not perturb NCP assembly, homogeneity or integrity at low ionic strength and are solvent-accessible with rotational freedom. Two essential features required for efficient FRET have been demonstrated: 1) The FL intensity of the Trp donors is independent of the state of NCP assembly; and 2) the FRET fluorophores, particularly the Cys-AEDANS acceptors, exhibit extensive rotational freedom.
H2A-H2B dimer dissociation monitored by FRET
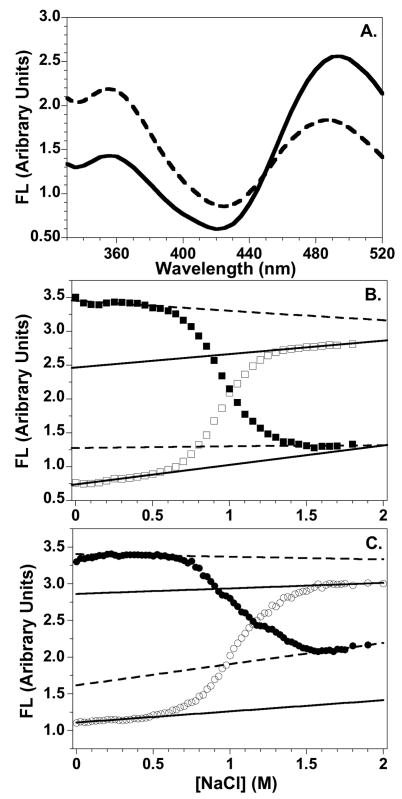
Representative emission scans of FRET NCPs at low and high NaCl concentrations are shown in Figure 5A. With excitation at 290 nm, the intact NCP exhibits quenched Trp FL and substantial Cys-AEDANS FL at 490 nm. Upon addition of sufficient NaCl to dissociate the H2A-H2B dimer, the Cys-AEDANS FL decreases, with a resulting increase in Trp FL at 350 nm. For the five NCP FRET pairs, decreases of 1.4 to 2.7 fold were observed for the Cys-AEDANS FL while the Trp FL increases by 1.7 to 3.8-fold. The range of FL changes correlates with the calculated Cβ donor-acceptor distances and whether the D-A and D-A′ distances are similar (as for the H2A Cys-AEDANS acceptors). The observed Cys-AEDANS FL at 1.5 M NaCl arises from an acceptor absorbance band around 275 nm,49 which yields some FL excitation at 490 nm, even upon excitation of the Trp donor at 290 nm. The FL intensity at 490 nm at 1.5 M NaCl in FRET NCPs is comparable to that observed for acceptor-only NCPs.
Figure 5.
Representative FL data for the salt-induced dissociation of the FRET NCPs. A. FL emission spectra of the H4-49W/H2A-108Cys-AEDANS FRET NCP at 0 M (solid line) and 1.5 M NaCl concentrations (dashed line). B and C. Salt dependence of the FL intensities for the H4-60W/H2A-108Cys-AEDANS and the H3-78W/H2B-109Cys-AEDANS NCPs, respectively. Trp donor FL at 350 nm: □ and ○; Cys-AEDANS FL at 490 nm: ■ and ●. Baselines for intact NCPs and the dimer-dissociated complex are indicated by solid and dashed lines, respectively. Conditions: 250 nM NCP; 20 mM Tris-Cl pH 7.6, 0.1 mM EDTA, 0.1 mM β-ME, 25°C.
The donor and acceptor FL intensities exhibit a sigmoidal dependence on [NaCl]. Representative transitions are shown in Figures 5B and 5C. Similar data were obtained for the other three NCP FRET pairs. The relatively steep, sigmoidal transitions observed by FRET are coincident with the first major transition detected by intrinsic Tyr FL of WT histones between 0.6 and 1.5 M NaCl (Figure 3). This coincidence strongly suggests that the FRET probes don’t alter NCP stability. The second transition observed by Tyr FL, above 1.5 M NaCl, reflects tetramer dissociation from DNA; as expected, the FRET signals are not sensitive to this dissociation step. Representative linear pre- and post-transition baselines are indicated in Figures 5B and 5C. The folded, pre-transition baselines were fitted using at least ten NaCl concentrations. The post-transition baselines were determined from at least five, and usually ten, NaCl concentrations. Between 0 and 0.2 M, the 490 nm FL of NCPs with the H2B-109Cys-AEDANS acceptor exhibited a small non-linear increase of ≤ 6% (Figure 5C). This curvature was not included in determination of the pre-transition baselines. No curvature was observed in the Trp FL baselines. The H2B-acceptor FL changes below 0.2 M NaCl may reflect local structural rearrangements or DNA breathing as observed in previous NCP FRET studies.15; 18
The salt-dependencies of the FRET data were analyzed by methods analogous to those established for protein folding studies. The assembled and disassembled baselines were used to convert the FL data to Fapp values (apparent fraction of the unfolded/dissociated species, Eq. 2, Methods section), allowing comparison of different FRET NCPs. These comparisons generally used Trp donor FL because the linearity of this signal from 0 to 0.5 M NaCl provided a less ambiguous determination of the pre-transition baselines. Figure 6 shows the Fapp values as a function of NaCl concentration for the five FRET NCPs. The three NCPs with the H2A-108Cys-AEDANS acceptor, but different Trp donors, are nearly coincident, with midpoints of ~0.95 M NaCl. The similarity in these transitions argues that the Trp donors, incorporated at different positions, have little effect on NCP stability.
Figure 6.
Fapp normalization of the NaCl-dependence of the Trp FL data for the FRET NCPs. Open symbols: NCPs with the H2A-108Cys-AEDANS acceptor and the H3-78W (○), H4-49W (◇), and H4-60W (□) donors. Closed symbols: NCPs with the H2B-109Cys-AEDANS acceptor and the H3-W78 (●) and H4-W60 (■) donors. Conditions are given in the legend of Figure 5.
Broader NaCl-induced transitions are observed for NCPs with the H2B acceptor than for NCPs with the H2A acceptor, despite common Trp donors. This acceptor-dependent difference in the steepness of the transitions may reflect differences in the D-A and D-A′ distances for H2A and H2B acceptors. These distances are similar for NCPs containing the H2A 108Cys-AEDANS acceptor. In contrast, ≥ 95% of the FRET signal for the H2B-109Cys-AEDANS NCPs arise from just one D-A pair. For the H2A-acceptor NCPs, both Trp donors are still within FRET distance of a potential acceptor (A or A′) in the hexameric species formed by dissociation of the first H2A-H2B dimer (Scheme 2). Thus, the donors in the hexameric population should be nearly homogeneous with respect to FRET signal. In contrast, half of the Trp donors are no longer quenched in the H2B-acceptor hexameric population. Two spectroscopically distinct donors in the hexameric species is akin to an unfolding intermediate, giving rise to a broader transition.50; 51 The small FL change of the H2B-Cys-AEDANS between 0.5 and 1.5 M NaCl may also contribute this heterogeneity and broadening of the transition region.
Free energy for the dissociation of the H2A-H2B dimer from the NCP
In this experimental system, the dissociation of the H2A-H2B dimers appears to be reversible. Samples were prepared at 1.0 to 1.2 M NaCl, where the dimer is 50% to 90% dissociated. Under these concentrations, Tyr FL indicates that the tetramer-DNA complex is still highly populated (Figure 3). These partially dissociated samples were diluted to 0.2 to 0.5 M NaCl by manual mixing and allowed to re-equilibrate for 10 min. The FL signals observed after refolding were within 5% of those of undissociated samples. This reversibility suggested that a thermodynamic free energy could be determined for the dissociation of the H2A-H2B dimers from the NCP.
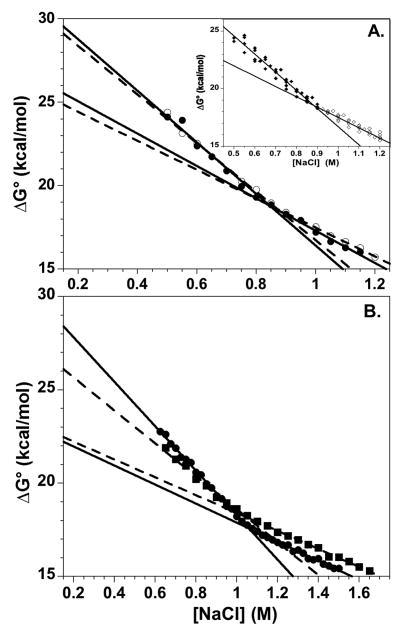
Thermodynamic analyses of transitions monitored by intrinsic Tyr FL and DNA-protein FRET15 are complicated by broad transitions that reflect contributions from the multiple steps: DNA breathing, dimer and tetramer dissociation. Thus, the relative concentrations of the various species can’t be accurately determined. An advantage of this protein-protein FRET system is that a single sigmoidal transition is observed which reports specifically on H2A-H2B dissociation. This allows determination of the relative populations of assembled and dissociated NCP species and estimation of equilibrium constant and free energy of dissociation at various NaCl concentrations (Eq. 3). Figure 7A shows a representative plot of the salt-dependent ΔG° values calculated for the transition region (0.6 to 1.3 M NaCl). The ΔG° values do not depend linearly on the NaCl concentration. The dependence of ΔG values for electrostatic interactions is often linear as a function of the square root of the ionic strength (for review52); this is not observed for the FRET NCP data (Supplementary material). However, the data for the five FRET NCPs were well described by two lines that intersect near the midpoint of the dissociation transition, 0.9 to 1 M NaCl. The two linear phases presumably reflect the dissociation of the first and then second H2A-H2B dimer. The fitted lines yield slopes which are analogous to the m values from protein folding studies, as well as estimates of the free energy of dissociation in the absence of NaCl, ΔG°(H2O) values (Eq. 1, Methods section). For the H2A-108Cys-AEDANS NCPs, with nearly coincident Fapp curves, the average slopes for the first and second transition regions were −15.8 ± 1.5 and −9.0 ± 0.8 kcal mol−1 M−1. The corresponding ΔG°(H2O) values were 32.5 ± 1.2 and 26.5 ± 0.8 kcal mol−1 at a 1 M standard state.
Figure 7.
The salt dependence of the free energy of dissociation for the H2A-H2B dimers from the NCP, showing extrapolations to physiological ionic strengths. A) H3-78W/H2A-108Cys-AEDANS NCP. Values were calculated from FL intensities at 350 nm (●, solid lines) and 490 nm (○, dashed lines) and were fit to two lines which intersect near the transition midpoint (Figure 6). Inset: overlay of data from the three H2A-108Cys-AEDANS NCPs; the lines represent the average ΔG°(H2O) and m values given in the text. B. H2B-109Cys-AEDANS NCPs with H3-78W (●, solid lines) or H4-60W (■, dashed lines). Conditions are given in legend of Figure 5.
The ΔG° values for NCPs with the H2B-109Cys-AEDANS are overlaid in Figure 7B. The free energies for the first transition are in good agreement, although the slopes differ by ~30% (11.9 and 8.8 kcal mol−1 M−1 with the H3 and H4 donors, respectively). In contrast, the slopes and ΔG° values for the second transitions differ by ≤ 6% (slopes of 5.1 and 4.8 kcal mol−1 M−1 for the H3 and H4 donors, respectively). Based on the slopes of the two transitions, dimer dissociation from the H2B acceptor NCPs appears to be less cooperative than from the H2A acceptor NCPs. This is consistent with a mixed population of hexameric species with differential donor quenching, as discussed above. When extrapolated to a physiological ionic strength of ~0.2 M, ΔG° values for the five FRET NCPs range between 25 and 29 kcal mol−1 (average = 28.6; standard deviation = 1.6) and 22 to 25 kcal mol−1 (average = 24.0; standard deviation = 1.4) for the first and second transitions, respectively.
FRET characterization of NCPs containing H2A.Z
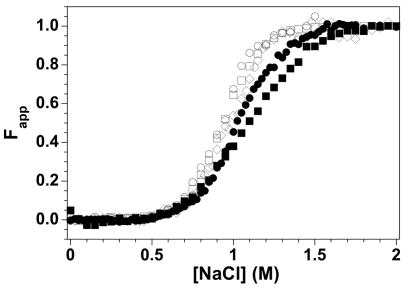
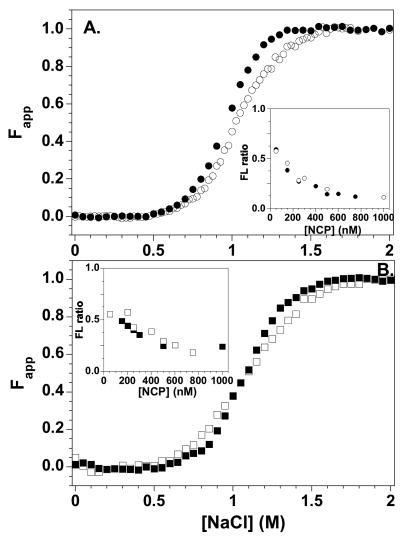
This multiple protein-protein FRET system is well-suited to characterize the effects of incorporation of histone variants on NCP equilibrium properties. H2A.Z was chosen because of contradictions in the literature regarding its biological function and effects on NCP stability. H2A.Z/H2B-109Cys-AEDANS dimers were reconstituted into NCPs with either H3-78W or H4-60W FRET donors. Native PAGE showed the quality and homogeneity of these NCPs were equivalent to those containing the major H2A (Figure 4B). Figure 8 shows a comparison of the salt-induced dissociation of H2A.Z and H2A NCPs. The transitions are fairly similar; the largest apparent difference is the steeper slope in the Fapp plots of the H2A.Z NCPs.
Figure 8.
Salt-induced equilibrium dissociation transitions for H2B-109Cys-AEDANS FRET NCPs with H2A (open symbols) and H2A.Z (solid symbols) normalized to Fapp values. A) H3-78W donor; B) H4-60W donor. The insets show the normalized ratios of the 350 nm and 490 nm FL as a function of NCP concentration at 1 M NaCl. Buffer conditions are given in the legend of Figure 5.
The stability of the NCPs were also assessed by altering the NCP concentration in 1.0 M NaCl, approximately the transition midpoint for the H2B acceptor NCPs (Figure 6). The ratios of the donor:acceptor FL as a function of NCP concentration are shown as insets in Figure 8. The FL ratio was used to compensate for the protein concentration dependence of the FL intensities, and normalized using Equation 2 (Methods) with YNCP set at the ratio for assembled NCP at 0.2 M and YT,D set at the ratio observed at 2.0 M NaCl (dimer fully dissociated from the NCP). Hence, a ratio of 0 is expected for the intact NCP, and the NCPs re-assemble at higher concentrations. Like the salt-dependent transitions, the FL ratios show very little difference in the stabilities of H2A and H2A.Z NCPs.
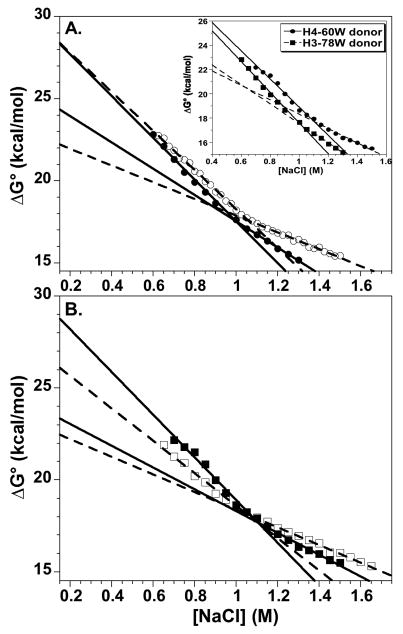
The salt-dependence of the ΔG° values for the H2A and H2A.Z NCPs with the H2B-109Cys-AEDANS acceptors are compared in Figure 9. As observed for H2A-containing NCPs, the ΔG° salt-dependencies are best described by two linear phases, which intersect near the transition midpoint. In the transition region, the ΔG° values for H2A and H2A.Z NCPs differ by ≤ 10%. Comparisons of the H2A and H2A.Z NCPs with the H3 and H4 FRET donors give slightly different results, but three generalities are apparent. 1) Extrapolations to physiological ionic strength suggest that dimer dissociation from the H2A.Z NCPs proceeds with an equal or somewhat greater ΔG° than from the H2A NCPs. 2) The H2A.Z NCPs with either Trp donor exhibit consistently lower ΔG° values for the second linear phase, i.e. dissociation of the second dimer is more favorable in H2A.Z NCPs, suggesting increased cooperativity. 3) The H2A.Z NCPs exhibit steeper slopes for both the first and second linear phases, 1.1 to 1.3-fold and 1.2 to 1.6-fold greater, respectively, which is also consistent with greater cooperativity in the disassembly of the H2A.Z NCP. In summary, although the stability differences between H2A and H2A.Z are small, there is an apparent increase in the cooperativity of dimer dissociation from the H2A.Z NCPs.
Figure 9.
Comparison of the salt dependence of the free energy of dissociation of the H2A-H2B dimers from H2A.Z and H2A NCPs, utilizing the H2B-109Cys-AEDANS acceptor. Linear extrapolations are shown to physiological ionic strengths. NCPs with the major H2A: open symbols, dashed lines; NCPs with the H2A.Z variant: solid symbols, solid lines. A) H3-78W donor. B) H4-60W donor. Inset panel A) Comparison of the two H2A.Z NCPs with H3-78W donor (circles) and H4-60W donor (squares). Conditions are given in legend of Figure 5.
DISCUSSION
Design of a protein-protein FRET system
An optimal FRET system employs solvent accessible fluorophores with substantial rotational freedom which do not significantly alter histone stability, and thus presumably histone or NCP structure. Comparison of the FL emission maxima and anisotropy r values for the folded and unfolded histones (Table 2) and the equilibrium stability of the modified histones (Figure 2), demonstrate that the engineered Trp and Cys-AEDANS residues exhibit these optimal properties. The native PAGE mobility of the variant NCPs (Figure 4) confirm that the FRET probes do not significantly alter NCP structure. Additional support for the absence of perturbation of NCP structure or stability is provided by the similarity of NaCl-induced transitions monitored by different spectral probes. The NaCl concentration range over which dissociation is observed in the five NCP FRET pairs overlap with the first transition observed for the WT 601-NCP monitored by Tyr Fl (0.6 to 1.5 M, Figures 3 and 6). The high similarity between FRET transitions with different Trp donors, especially with the H2A-108Cys-AEDANS acceptor, is also consistent with the FRET probes causing minimal changes in NCP structure or stability.
The salt-dependent loss of the FRET signal has been attributed to dissociation of the H2A-H2B dimers. It is conceivable that the FRET signal arises instead from a conformational change which increases the distances between the dimers and tetramer to substantially greater than the 20 Å Ro, without dimer dissociation. However, three lines of evidence argue strongly against this interpretation. Firstly, the FRET signal in the middle of the transition exhibits substantial protein concentration dependence as expected for a dissociation reaction (insets of Figure 8). Secondly, the transitions monitored by FRET overlap with that monitored by Tyr FL, which has been well-established as arising from H2A-H2B dissociation.53 Thirdly, probes which monitor distances between a range of intra-nucleosomal positions yield overlapping FRET transitions. It is difficult to conceive of a conformational change which would similarly increase all of these distances but not involve dissociation.
To determine a free energy, ΔG°, for the dissociation of the H2A-H2B dimers from the NCP, the signal change must monitor a single transition, clearly separated from other reactions (such as DNA breathing or tetramer dissociation), and the reaction must be reversible. The FL of NCPs containing only Trp donors or Cys-AEDANS acceptors is largely insensitive to dissociation events. In contrast, the salt dependencies of the donor and acceptor FL signals in the FRET NCPs are sigmoidal, indicative of a single, cooperative transition corresponding to the dissociation of the two H2A-H2B dimers. These data can then be used to determine the relative populations of NCP and [free dimers + tetramer-DNA complex]. To accurately determine the populations of different species from spectral changes, well-defined pre- and post-transition baselines are required—which allows estimation of the signals expected for fully assembled and disassembled species throughout the transition region. The greater stability of NCPs reconstituted with the 601 positioning sequence and the insensitivity of FRET to multiple transitions made this possible, in contrast to previous FRET systems.15; 18 The criteria of reversibility was demonstrated by the ~95% recovery of the expected FRET signals in samples refolded by dilution from salt concentrations at which the majority of the H2A-H2B dimers have dissociated.
Comparisons to previously reported NCP FRET systems
FRET systems have been described previously for the study of NCP dynamics,15; 18–20 monitoring DNA-DNA, protein-DNA and protein-protein interactions, with a focus on DNA movements and usually at least one of the FRET probes attached to the DNA. There are three significant advantages or distinctions regarding the Trp/Cys-AEDANS FRET system described here: 1) relatively small size and hydrophobic surface area of the D-A pair; 2) a 20 Å Förster distance that is substantially smaller than the dimensions of the NCP and ability to specifically monitor the changes in distance between the H2A-H2B dimers and the (H3-H4)2 tetramer; and 3) the use of the 601 NCP positioning sequence.
The D-A pairs used in previous studies were larger and more hydrophobic: coumarin/fluorescein15 and Cy3/Cy5.18; 19; 39; 40 Such hydrophobic dyes have a tendency to sequester themselves from solvent,54 which can perturb macromolecular structures. This may be a concern in the coumarin/fluorescein FRET system,15 given the large increase in anisotropy of the fluorophores in the NCP compared to the free fluorophores (~0.19 and 0.05, respectively). No report of the anisotropy data of the Cy3/Cy5 fluorophores is apparent. Additionally, the larger Förster distances of previously used D-A pairs, 50 to 60 Å, are comparable to the molecular dimensions of the NCP (100 Å in diameter by 50 Å in width). Under physiological conditions, H2A-H2B dimers are able to undergo rapid exchange between partially assembled NCPs and form unstable DNA/H2A-H2B complexes.55; 56 Above 0.5 M NaCl, H2A-H2B dimers may weakly associate with DNA in the DNA-tetramer complexes in a non-NCP conformation that would be detectable by FRET probes with R0 values of ~50Å. This complexity is suggested by the difference in midpoints attributed to dimer dissociation from 5S-rRNA NCPs determined from DNA-DNA (~0.65 M), DNA-H4 (~0.8 M) and H4-H2B FRET pairs (~0.6 M).15 With a shorter R0, detection of non-specific interactions should be minimized. Conversely, a shorter R0 may be more sensitive to local conformational changes, leading to FRET changes that do not reflect dimer dissociation; however, consistent results between multiple protein-protein FRET pairs suggets that this potential complication is not an issue.
A third important difference between the current and previous FRET systems15; 20 is the use of the 601 NCP positioning sequence. This sequence yields a more stable, homogeneously positioned nucleosome38 than the historically used 5S rRNA gene sequence and is becoming a standard positioning sequence in chromatin studies (e.g.18; 19; 57; 58). There are reports on the kinetics of DNA breathing in the context of the 601 sequence,18; 19 but no biophysical studies describing effects on histone dissociation. This is the first report characterizing the salt-induced dissociation of the dimer from 601-NCPs. Interestingly, it is the dimer that is stabilized by the 601 sequence, with little to no effect on the dissociation of the tetramer, despite the central importance of the tetramer in NCP positioning.59 0.1 to 0.2 M higher NaCl concentrations are required to initiate dissociation of the H2A-H2B dimers from 601 NCPs, compared to the 5S-rRNA NCPs. In contrast, the Tyr Fl data show that there is little effect on the initiation of the (H3-H4)2 tetramer dissociation at 1.5 M NaCl. The effect of DNA sequence on H2A-H2B affinity for the NCP has been noted recently in a DNA-DNA FRET study.39 These results have important biological implications because of the array of complexes which promote dissociation and/or exchange of H2A-H2B dimers from the NCP.60
Free energy of H2A-H2B dimer dissociation
Unique aspects of this FRET system (well-defined, reversible sigmoidal transition, discussed above) allow the calculation of a free energy for dimer dissociation. Previous thermodynamic measurements have been limited to relative binding free energies, ΔΔG, between various DNA sequences, using competitive gel shift assays (for review61). Dilution-driven dissociation experiments to determine an absolute equilibrium constant for nucleosome disassembly have been unsuccessful.56; 62 The dissociation reactions were not reproducible unless destabilizing additives were used to prevent nonspecific loss of dissociated histones; even with additives, dilution-driven dissociation was not fully reversible, preventing the determination of histone-DNA binding free energies. The dilution driven experiments also demonstrated that a simple dissociation model (NCP ⇆ free DNA + histone octamer) is inappropriate, and that multiple non-nucleosomal histone-DNA species must be considered.56 Therefore, a first step toward a thermodynamic description of NCP assembly should begin with analysis of the ΔG° for dimer dissociation, for which the current FRET system is ideally suited. Salt-induced dissociation, shown to be reversible, was employed instead of dilution-driven dissociation. However, analyses of perturbation experiments do require extrapolation to physiological ionic strengths.
Interestingly, two transitions were evident in the salt-dependence of the ΔG° values for all FRET NCP combinations (Figures 7 and 9). This biphasic response is interpreted as dissociation of the first and then second H2A-H2B dimer, with different affinities for the tetramer-DNA complex. The population of hexameric NCP intermediates have been reported in the presence of histone chaperones63 and depletion of chicken erythrocyte core particles under certain conditions.64 The presence of cooperativity in the dissociation of the H2A-H2B dimers has been suggested previously, but not demonstrated quantitatively. In the current study, the extrapolated ΔG° values for the second transition are less than for the first, indicating positive cooperativity. The second dimer dissociates more easily than the first, with a ΔG° that is ~20% lower at physiological ionic strength. The close proximity of the H2A L1 loops in the NCP has been proposed to play a role in the cooperative binding of the H2A/H2B dimers.8; 10; 65
The data in this report appear to be one of the first determinations of a free energy for dimer dissociation using a well-defined, homogeneous mono-nucleosome system. A ΔG of −15.5 kcal mol−1 was reported for association of two H2A-H2B dimers with the H3-H4 tetramer to form the histone octamer in 2 M NaCl where the octamer is stable in the absence of DNA.53 The apparent dissociation constant for H2A-H2B dimers from chicken erythrocyte long chromatin yielded a ΔG of ~9 kcal/mol in 1 M NaCl,14 compared to 17 to 18 kcal/mol determined for the 601-NCP FRET system (Figure 7). Part of this difference can be attributed to the known differential affinity of the histone octamer for random chicken erythrocyte DNA and 601 DNA, ΔΔG = 3.5 kcal mol−1.61 The standard state concentration for the study employing long chromatin is also unclear.
Thermodynamic comparison of H2A and H2A.Z containing NCPs
The structural and functional consequences of incorporating the H2A.Z variant into NCPs have been controversial in the literature (for review,33). Comparison of the crystal structures of H2A and H2A.Z NCPs suggested that some interactions may stabilize H2A.Z NCPs, while others may be destabilizing.10 Initially, Ausio and colleagues reported a significant destabilization of H2A.Z NCPs based on salt-dependent analytical ultracentrifugation studies.34 This study used chicken erythrocyte histones and recombinant human H2A.1 or H2A.Z to form NCPs with random 146 bp DNA fragments or 12mer oligonucleosome arrays with the 208 bp sea urchin 5S DNA. A subsequent FRET study using recombinant Xenopus laevis histones, recombinant mouse H2A.Z and the sea urchin 5S DNA suggested an increase in stability for the H2A.Z NCPs.15 Three FRET pairs monitoring DNA-DNA, DNA-H2B and H2B-H4 interactions showed an apparent shift to higher NaCl concentrations for dissociation of the H2A.Z/H2B dimers. However, the ΔCM values of 36 to 66 mM are close to the experimental uncertainty of the experiments. Likewise, the slopes for the dissociation transitions were similar or slightly smaller for the H2A.Z NCPs, suggesting a decrease in cooperativity or a difference in the salt sensitivities of the H2A and H2A.Z molecular interactions in the NCP. However, the ΔCM values and slopes may be influenced by the choice of baselines during normalization, which is more ambiguous in such mutliphasic transitions, unlike the current study. A recent study by Ausio and colleagues35 contradicted their previous report, attributing the differences to the histone source and purification methods. This later study used histones isolated exclusively from chicken erythrocytes. Sedimentation analytical ultracentrifugation and separation of nucleosomal components by sucrose gradients or hydroxyapatite chromatography were used to analyze salt dependent NCP dissociation. The H2A.Z NCPs exhibited a subtle increase in stability, which was abolished by histone acetylation. It is apparent from the conflicting literature reports that the context of the NCPs, i.e. the source of the histones, their modification state and the DNA sequence used, is an important consideration.
The results presented here confirm that the effects of H2A.Z on NCP stability are small and subtle. The H2A and H2A.Z NCP dissociation transitions have similar midpoints (Figure 8), although the slopes in plots of Fapp and ΔG° (Figures 8 and 9) are somewhat steeper for H2A.Z NCPs, suggesting greater cooperativity in dimer dissociation relative to H2A NCPs. Enhanced cooperativity may reflect changes in the interactions between the L1 loops of H2A and H2A′. More hydrogen bonds and contacts between the L1-L1′ loops are observed in the H2A.Z NCP structure compared to the H2A NCP structure.10 These differences were predicted to cause more cooperativity in the dissociation of the two dimers, which is the major effect observed in the current study. In contrast, fewer hydrogen bonds in the H2A.Z-H3 interface were predicted to be destabilizing, but appear not to be a major determinant of stability for H2A and H2A.Z NCPs, given the results of this and other studies15; 35 which are consistent with marginally greater stability for H2A.Z NCPs.
The increased cooperativity observed in this study is in opposition to the apparent decreased cooperativity observed in the previous FRET study.15 Similar recombinant histones were used, but were complexed with different DNAs (the artificial 601 sequence here instead of the 5S gene sequence). Decreased cooperativity in the previous study might reflect non-NCP interactions of the H2A(.Z)/H2B dimers after dissociation, detected by virtue of the larger Förster distance of the coumarin/fluorescein FRET pair. Alternatively, the increased cooperativity observed in the current study may reflect the greater stability of NCPs containing the 601 DNA. Although it is believed that positioning effects result primarily from tetramer-DNA interactions,59 the results of this study demonstrate that the 601 sequence can enhance the affinity of the H2A-H2B dimers for tetramer-DNA complexes. The extent to which the 601 DNA sequence dictates the stability and cooperativity of dimer dissociation can be easily determined with this protein-protein FRET system, using different DNA sequences.
Implications and future applications of a protein-protein FRET system
The nucleosome core particle is critical in the compaction of chromatin and regulation of DNA-templated chemistries. Significant chromatin remodeling must take place in order for the diverse machinery of replication, transcription, recombination and repair access nucleosomal DNA. In addition to the inherent mobility of the nucleosome, the cell employs various mechanisms such as ATP-dependent chromatin remodeling complexes, histone chaperones, histone variants, and post-translation modifications to modulate DNA access.3; 66 The protein-protein FRET system described herein is well-suited for dissection of the kinetic processes of dimer association and dissociation in the assembly and disassembly of the NCP. The utility of this FRET system to determine the effects of histone variants on NCP dynamics has also been demonstrated. It may also be amenable to the study of catalytic amounts of chromatin remodeling complexes or histone chaperones. Overall, protein-protein FRET is a powerful and versatile tool for elucidating the relationships between histone sequence and stability, DNA sequence and NCP dynamics.
MATERIALS AND METHODS
Materials
The following chemicals were purchased from the indicated companies: ultrapure urea, MP Biomedicals (Solon, OH); ultrapure GdmCl, EMD Chemicals (Gibbstown, NJ); 1,5-IAEDANS, Invitrogen (Carlsbad, CA); and trimethylamine N-oxide (TMAO), Sigma (St. Louis, MO) and prepared as described previously.43 All other chemicals were of reagent grade. The oligonucleotides used for mutagenesis and the construction of pLMG601-23 were purchased from MWG-BIOTECH Inc. (High Point, NC).
Methods
Sample preparation
Histone mutagenesis, expression and purification
The T7 pET expression plasmids containing the Xenopus laevis histone genes47 were used as templates for site-directed mutagenesis. QuikChange® II Mutagenesis Kits (Stratagene, La Jolla, CA) were used to generate the following mutations: H2A-L108C, H2B-S109C, H3-F78W, H4-L49W and H4-V60W. The presence of the desired mutation and absence of other mutations were confirmed by sequencing the entire histone gene. The recombinant histones were overexpressed in the BL21(DE3)pLys E. coli strain and purified as described previously.43; 67 H2A.Z was overexpressed from a pET vector containing the mouse gene10 and purified by methods described previously.68 SDS-PAGE analyses showed that the histone preparations were ≥ 90% pure (histone and NCP reconstitution provided further purification). Purified monomers were dialyzed against 10 mM HCl, 3 mM β-mercaptoethanol; H3 and H4 were then lyophilized.
Histone labeling
H2A and H2B Cys mutant monomers were modified with the thiol-specific reagent 1,5-IAEDANS in a buffer of 6 M urea, 50 mM KCl and 20 mM Tris-Cl, pH 7.5 without any reducing agent. 4 mg/ml IAEDANS, in 20 mM Tris-Cl pH 7.5 was added slowly with stirring to a final molar ratio of 20:1 IAEDANS to Cys. The labeling reaction was incubated for 3 hours at room temperature (under vacuum). The reaction was quenched with a 10 fold excess of β-ME followed by exhaustive dialysis into a buffer of 6 M urea, 0.2 M KCl, 0.1 mM K2EDTA, 3 mM β-ME and 20 mM KPi, pH 7.2. The Cys-AEDANS content of the monomers was calculated using an extinction coefficient of 5,700 M−1cm−1 at 336 nm, and compared to the protein concentration determined by BCA assays (reagents from Sigma, St. Louis, MO). The extent of labeling was ≥90%, which was also confirmed by triton/acid/urea gels, which can resolve histone post-translational modifications.69
Histone reconstitution
Purified recombinant histones were refolded to their appropriate hetero-oligomeric form by methods similar to those described previously.43; 67 Urea-unfolded H2A and H2B monomers were combined in a 1:1 ratio and refolded by 5-fold dilution into buffer without urea, and dialyzed to remove remaining urea. The refolded H2A-H2B dimers were further purified with a Heparin column as described previously.67 Lyophilized H3 and H4 monomers were dissolved in a buffer of 8 M GdmCl, 5 mM K2EDTA, 5 mM β-ME and 20 mM KPi, pH 7.4, and then diluted 10-fold into buffer without denaturant. The reconstituted H3-H4 preparation was dialyzed to remove residual GdmCl and then concentrated in the dialysis tubing by placing the tubing in solid PEG 10,000, followed by further dialysis. The concentrations of the folded WT and Trp mutant oligomers were determined in 6 M GdmCl buffer using extinction coefficients calculated by the method of Gill and von Hippel.70 The Cys-AEDANS H2A-H2B dimer concentrations were determined using the BCA assay because of the contribution of Cys-AEDANS to absorbance at 280 nm.
601 DNA production
NCPs were reconstituted with a 149 bp segment of the unnatural 601 sequence.37; 38 The central 149 bp of the 601 sequence was amplified by PCR from the pGEM3Z plasmid, kindly provided by the lab of Dr. Jonathan Widom (Northwestern University). To enhance DNA production, a pUC119-based plasmid, pLMG601-23, was constructed which contained 23 tandem repeats of the 149 bp of 601 sequence flanked by EcoRV restriction sites. The pLMG601-23 plasmid was produced and extracted from E. coli DH5α using a modified alkali lysis protocol described previously.71 149 bp segments of 601 DNA were released by EcoRV digestion and isolated with differential precipitation using PEG 6000.71 The quality of the digestion and isolation was monitored by 5% native PAGE. The DNA concentration was determined by absorbance at 260 nm.
Octamer and NCP formation
The appropriate ratio of folded H2A-H2B dimers and H3-H4 tetramers were combined in a buffer of 2 M KCl, 5 mM Na2EDTA, 5 mM β-ME and 20 mM KPi, pH 7.45, and dialyzed against this buffer to allow formation of histone octamers. After concentration with a Centricon YM-10 (Millipore Inc.; Billerica, MA), the octamer preparation was purified by size-exclusion chromatography with a BIOSEP SEC-3000 HPLC column, equilibrated in the 2 M KCl buffer described above, which removed improperly assembled oligomers. Fractions were analyzed by 15 % SDS-PAGE, and those with the appropriate histone ratio of 1:1:2 for H3, H4 and H2A-H2B (which co-migrate) were pooled.
NCP reconstitution was achieved by combining DNA and histone octamers in the 2 M KCl buffer and lowering the ionic strength by step dialysis against buffers with 0.85, 0.6, 0.2 and 0 M KCl. The histone octamer concentration was estimated from the Cys-AEDANS absorbance at 336 nm. Since the labeling efficiency was not 100%, the octamer concentration was slightly inaccurate. Therefore, the optimal DNA:octamer ratio for NCP reconstitution was determined from 50 μl reactions with DNA and octamer ratios varied from 0.4:1 to 1.2:1, using mini-buttons for dialysis.61 5% native PAGE assessed reconstitution quality and identify the DNA:octamer ratio which yielded the greatest amount of NCP and minimal free DNA, typically a ratio of 0.8:1. This ratio was then used in the large-scale reconstitutions (~0.6 mg/ml NCP in 4 to 6 ml) with conventional step dialysis methods, with a final dialysis step into the buffer used in spectroscopy experiments (20 mM Tris, pH 7.6, 1 mM EDTA and 1 mM β-ME). After dialysis, the reconstituted NCPs were centrifuged at 16.1 x g at 25 °C for 5 min to remove any aggregates.
Data collection and analyses
Instrumentation
Spectroscopy data were collected on an AVIV 202SF circular dichroism spectrophotometer and an AVIV ATF-105/305 differential/ratio spectrofluorometer. Both instruments were interfaced with Hamilton Model 500 automated titrators, used for collecting equilibrium stability data. The spectrometers were equipped with peltier-controlled sample holders; all spectroscopic data were collected at 25 °C.
Equilibrium stability studies of the H2A-H2B dimer and H3-H4 tetramer
Protein unfolding titrations were monitored by far-UV CD (222 nm) and FL, with 2 min equilibrations at each denaturant concentration. The equilibration times are significantly longer than the slowest observed folding and unfolding kinetic phases for the oligomers.68; 72 For H2A-H2B mutants, intrinsic Tyr FL emission at 305 nm was used to monitor quaternary and tertiary unfolding, with excitation at 280 nm. H3-H4 unfolding was monitored by Trp FL emission at 340 nm, with excitation at 290 nm.
The buffer conditions (20 mM KPi, pH 7.2; 0.2 M KCl, 0.1 mM EDTA) were the same as in previous studies with WT recombinant histones.43; 67 Urea was used as the denaturant in the H2A-H2B equilibrium titrations. H3-H4 titrations employed GdmCl as the denaturant to improve the reversibility of tetramer unfolding.43 The buffers also contained 1 M TMAO to stabilize the tetramer and yield a well-defined native baseline.
Thermodynamic parameters were determined from the equilibrium data by fits to the well-established linear extrapolation method,73; 74 described in Equation 1:
| (1) |
where ΔG°(H2O) is the free energy of unfolding in the absence of denaturant at a 1 M standard state and the m value describes the sensitivity of the transition to denaturant and correlates to the change in solvent accessible surface area upon unfolding.75 The global fitting program Savuka 5.176; 77 was used to globally fit data collected at varied protein concentrations and with different spectral probes. The error at one standard deviation of the fitted parameters were estimated using a rigorous analyses of the error surface as described elsewhere.78
FL anisotropy of modified histones oligomers
The FL anisotropy of the Cys-AEDANS and Trp residues was measured for the folded and denaturant-unfolded histone oligomers and the assembled NCPs. The G-factor (G), the ratio of the sensitivity of the instrument monochromaters for vertically and horizontally polarized light,42 was determined for each emission wavelength.
Steady-state FL and FRET in the NCPs
A buffer of 20 mM Tris-Cl, pH 7.6, 1 mM EDTA and 1 mM β-ME was used in all NCP studies. The NCP concentration in the salt-induced dissociation transitions was 250 nM, comparable to a previous NCP FRET study.15 FL excitation was at 290 nm, and emission was monitored at 350 and 490 nm for Trp and Cys-AEDANS, respectively. A relatively narrow excitation bandwidth of 1 nm was employed to minimize photobleaching, with an emission bandwidth of 4 nm. The salt concentration was varied by manual titration of NCP samples with buffer containing 5 M NaCl. Over the course of a titration, the NaCl concentration changed by ~0.5 M, while the NCP concentration varied less than 10%. Samples were equilibrated for 5 minutes after each incremental salt increase of 0.025 to 0.05 M NaCl before the FL intensities were recorded. Data points in Figures 5, 6 and 8 represent the average of two or three measurements from titrations initiated at different salt concentrations.
The FL of the Trp donor (350 nm) and Cys-AEDANS acceptor (490 nm) were normalized to Fapp, the apparent fraction of NCP dissociated to H2A-H2B dimers and tetramer-DNA complex (T·DNA), as defined by Equation 2:
| (2) |
where [NCPinitial] is the initial concentration of folded NCP, Yi is the FL at a given [NaCl]; YNCP describes the linear response of the FL signal of the intact NCP (i.e. the folded baseline) and YT,D is the linear response observed for the disassembled population of the H3-H4/DNA complex and the free H2A-H2B dimers.
Since a single, smooth transition is observed, the free energy of dimer dissociation can only be calculated for the overall conversion of NCP to T·DNA + 2D (Scheme 1), rather than a free energy for the dissociation of each dimer (Scheme 2). The overall ΔG° is determined by Keq = K1·K2, where K1 and K2 represent the equilibrium constants for dissociation of the first and second dimers, respectively. From the Fapp values in the transition region (0.5 to 1.3 M NaCl), the concentrations of the NCP, H2A-H2B dimer and tetramer-DNA complex were determined and used to calculate KEq according to Equation 3:
| (3) |
given the following relationships: [T·DNA] = Fapp[NCPinitial]; [D] = 2[T·DNA]; and [NCP] = [NCPinitial] – [T·DNA].
Supplementary Material
Acknowledgments
This work was supported by an N.I.H. grant (GM073787) to LMG. DAH was supported in part by an N.I.H. Biotechnology pre-doctoral training grant (T-32 GM008336). The 601 nucleosome positioning sequence was kindly provided by the lab of Dr. Jonathan Widom (Northwestern University). The expression plasmids for the WT major Xenopus laevis histones and the H2A.Z variant from Mus muculus were provided by Dr. Karolin Luger (Colorado State University).
Footnotes
Abbreviations: β-ME, beta-mercaptoethanol; CD, circular dichroism; CM, midpoint of equilibrium titration curve; Cys-AEDANS, Cys residues modified with 1,5-IAEDANS; EtBr, ethidium bromide; Fapp, apparent fraction dissociated; FL, fluorescence; FRET, Förster resonance energy transfer; ΔG°(H2O), the free energy of unfolding in the absence of denaturant; GdmCl, guanidinium chloride; 1, 5-IAEDANS, 5-((((2-iodoacetyl)amino)ethyl)-amino)-naphthalene-1-sulfonic acid; KPi, potassium phosphate, pH 7.2; m value, parameter describing the sensitivity of the unfolding transition to the [denaturant]; NCP, nucleosome core particle; PAGE, polyacrylamide gel electrophoresis; PEG, polyethylene glycol; TMAO, trimethylamine-N-oxide; WT, wild-type
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Ann Rev Biochem. 1998;67:545–79. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–72. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 3.Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. doi: 10.1007/s10577-005-1026-1. [DOI] [PubMed] [Google Scholar]
- 4.Wolffe AP. Chromatin remodeling: why it is important in cancer. Oncogene. 2001;20:2988–90. doi: 10.1038/sj.onc.1204322. [DOI] [PubMed] [Google Scholar]
- 5.Huang C, Sloan EA, Boerkoel CF. Chromatin remodeling and human disease. Curr Opin Genet Dev. 2003;13:246–52. doi: 10.1016/s0959-437x(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 6.Yasui W, Oue N, Ono S, Mitani Y, Ito R, Nakayama H. Histone acetylation and gastrointestinal carcinogenesis. Ann N Y Acad Sci. 2003;983:220–31. doi: 10.1111/j.1749-6632.2003.tb05977.x. [DOI] [PubMed] [Google Scholar]
- 7.Arents G, Moudrianakis EN. The histone fold: A ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc Natl Acad Sci USA. 1995;92:11170–74. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 9.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 10.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–4. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 11.Oohara I, Wada A. Spectroscopic studies on histone-DNA interactions. I The interaction of Histone (H2A,H2B) dimer with DNA: DNA sequence dependence. J Mol Biol. 1987;196:389–97. doi: 10.1016/0022-2836(87)90699-1. [DOI] [PubMed] [Google Scholar]
- 12.Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–80. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akey CW, Luger K. Histone chaperones and nucleosome assembly. Curr Opin Struct Biol. 2003;13:6–14. doi: 10.1016/s0959-440x(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 14.Oohara I, Wada A. Spectroscopic studies on histone-DNA interactions. II Three transitions in nucleosomes resolved by salt-titration. J Mol Biol. 1987;196:399–411. doi: 10.1016/0022-2836(87)90700-5. [DOI] [PubMed] [Google Scholar]
- 15.Park YJ, Dyer PN, Tremethick DJ, Luger K. A new FRET approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J Biol Chem. 2004;13:24274–82. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 16.Polach KJ, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol. 1996;258:800–12. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- 17.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–49. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 18.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat Struct Mol Biol. 2004;11:763–9. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 19.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 20.Tomschik M, Zheng H, van Holde K, Zlatanova J, Leuba SH. Fast, long-range, reversible conformational fluctuations in nucleosomes revealed by single-pair fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 2005;102:3278–83. doi: 10.1073/pnas.0500189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ausio J, Abbott DW, Wang X, Moore SC. Histone variants and histone modifications: a structural perspective. Biochem Cell Biol. 2001;79:693–708. [PubMed] [Google Scholar]
- 22.Ausio J, Abbott DW. The many tales of a tail: carboxyl-terminal tail heterogeneity specializes histone H2A variants for defined chromatin function. Biochemistry. 2002;41:5945–9. doi: 10.1021/bi020059d. [DOI] [PubMed] [Google Scholar]
- 23.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162–9. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 24.Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005;21:133–53. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- 25.Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 26.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–36. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 27.Rangasamy D, Berven L, Ridgway P, Tremethick DJ. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. Embo J. 2003;22:1599–607. doi: 10.1093/emboj/cdg160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol Cell. 2004;16:655–61. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 29.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–48. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–31. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dryhurst D, Thambirajah AA, Ausio J. New twists on H2A.Z: a histone variant with a controversial structural and functional past. Biochem Cell Biol. 2004;82:490–7. doi: 10.1139/o04-043. [DOI] [PubMed] [Google Scholar]
- 33.Raisner RM, Madhani HD. Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr Opin Genet Dev. 2006;16:119–24. doi: 10.1016/j.gde.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausio J. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J Biol Chem. 2001;276:41945–9. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- 35.Thambirajah AA, Dryhurst D, Ishibashi T, Li A, Maffey AH, Ausio J. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J Biol Chem. 2006;281:20036–44. doi: 10.1074/jbc.M601975200. [DOI] [PubMed] [Google Scholar]
- 36.Flaus A, Rencurel C, Ferreira H, Wiechens N, Owen-Hughes T. Sin mutations alter inherent nucleosome mobility. Embo J. 2004;23:343–53. doi: 10.1038/sj.emboj.7600047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 38.Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol. 1999;288:213–29. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 39.Kelbauskas L, Chan N, Bash R, Yodh J, Woodbury N, Lohr D. Sequence-dependent nucleosome structure and stability variations detected by Forster resonance energy transfer. Biochemistry. 2007;46:2239–48. doi: 10.1021/bi061289l. [DOI] [PubMed] [Google Scholar]
- 40.Lovullo D, Daniel D, Yodh J, Lohr D, Woodbury NW. A fluorescence resonance energy transfer-based probe to monitor nucleosome structure. Anal Biochem. 2005;341:165–72. doi: 10.1016/j.ab.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 41.Dalbey RE, Weiel J, Yount RG. Forster energy transfer measurements of thiol 1 to thiol 2 distances in myosin subfragment 1. Biochemistry. 1983;22:4696–706. doi: 10.1021/bi00289a014. [DOI] [PubMed] [Google Scholar]
- 42.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2. Plenum Press; New York: 1999. [Google Scholar]
- 43.Banks DD, Gloss LM. Equilibrium folding of the core histones: the H3-H4 tetramer is less stable than the H2A-H2B dimer. Biochemistry. 2003;42:6827–39. doi: 10.1021/bi026957r. [DOI] [PubMed] [Google Scholar]
- 44.Wang A, Bolen DW. A naturally occurring protective system in urea-rich cells: mechanism of osmolyte protection of proteins against urea denaturation. Biochemistry. 1997;36:9101–8. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- 45.Bolen DW. Protein stabilization by naturally occurring osmolytes. Methods Mol Biol. 2001;168:17–36. doi: 10.1385/1-59259-193-0:017. [DOI] [PubMed] [Google Scholar]
- 46.Henkels CH, Kurz JC, Fierke CA, Oas TG. Linked folding and anion binding of the Bacillus subtilis ribonuclease P protein. Biochemistry. 2001;40:2777–89. doi: 10.1021/bi002078y. [DOI] [PubMed] [Google Scholar]
- 47.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–11. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 48.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 49.Steer BA, Merrill AR. The colicin E1 insertion-competent state: detection of structural changes using fluorescence resonance energy transfer. Biochemistry. 1994;33:1108–15. doi: 10.1021/bi00171a009. [DOI] [PubMed] [Google Scholar]
- 50.Spudich G, Marqusee S. A change in the apparent m value reveals a populated intermediate under equilibrium conditions in Escherichia coli ribonuclease HI. Biochemistry. 2000;39:11677–83. doi: 10.1021/bi000466u. [DOI] [PubMed] [Google Scholar]
- 51.Hobart SA, Meinhold DW, Osuna R, Colon W. From two-state to three-state: The effect of the P61A mutation on the dynamics and stability of the Factor for Inversion Stimulation in an altered equilibrium denaturation mechanism. Biochemistry. 2002;41:13744–54. doi: 10.1021/bi0265224. [DOI] [PubMed] [Google Scholar]
- 52.Record MT, Jr, Zhang W, Anderson CF. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: A practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects and osmotic effects of salts. Advances in Protein Chemistry. 1998;51:282–355. doi: 10.1016/s0065-3233(08)60655-5. [DOI] [PubMed] [Google Scholar]
- 53.van Holde K. Chromatin. In: Rich A, editor. Springer Series in Molecular Biology. Springer-Verlag; New York: 1989. [Google Scholar]
- 54.Randolph JB, Waggoner AS. Stability, specificity and fluorescence brightness of multiply-labeled fluorescent DNA probes. Nucleic Acids Res. 1997;25:2923–9. doi: 10.1093/nar/25.14.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aragay AM, Diaz P, Daban JR. Association of nucleosome core particle DNA with different histone oligomers: Transfer of histones between DNA-(H2A,H2B) and DNA-(H3,H4) complexes. J Mol Biol. 1988;204:141–54. doi: 10.1016/0022-2836(88)90605-5. [DOI] [PubMed] [Google Scholar]
- 56.Thastrom A, Gottesfeld JM, Luger K, Widom J. Histone-DNA binding free energy cannot be measured in dilution-driven dissociation experiments. Biochemistry. 2004;43:736–41. doi: 10.1021/bi0302043. [DOI] [PubMed] [Google Scholar]
- 57.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–3. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 58.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–41. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 59.Dong F, van Holde KE. Nucleosome positioning is determined by the (H3-H4)2 tetramer. Proc Natl Acad Sci USA. 1991;88:10596–600. doi: 10.1073/pnas.88.23.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thiriet C, Hayes JJ. Histone dynamics during transcription: exchange of H2A/H2B dimers and H3/H4 tetramers during pol II elongation. Results Probl Cell Differ. 2006;41:77–90. doi: 10.1007/400_009. [DOI] [PubMed] [Google Scholar]
- 61.Thastrom A, Lowary PT, Widom J. Measurement of histone-DNA interaction free energy in nucleosomes. Methods. 2004;33:33–44. doi: 10.1016/j.ymeth.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 62.Gottesfeld JM, Luger K. Energetics and affinity of the histone octamer for defined DNA sequences. Biochemistry. 2001;40:10927–33. doi: 10.1021/bi0109966. [DOI] [PubMed] [Google Scholar]
- 63.Mazurkiewicz J, Kepert JF, Rippe K. On the mechanism of nucleosome assembly by histone chaperone NAP1. J Biol Chem. 2006;281:16462–72. doi: 10.1074/jbc.M511619200. [DOI] [PubMed] [Google Scholar]
- 64.Read CM, Baldwin JP, Crane-Robinson C. Structure of subnucleosomal particles. Tetrameric (H3/H4)2 146 base pair DNA and hexameric (H3/H4)2(H2A/H2B)1 146 base pair DNA complexes. Biochemistry. 1985;24:4435–50. doi: 10.1021/bi00337a027. [DOI] [PubMed] [Google Scholar]
- 65.Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–52. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luger K. Structure and dynamic behavior of nucleosomes. Curr Opin Genet Dev. 2003;13:127–35. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 67.Gloss LM, Placek BJ. The Effect of Salts on the Stability of the H2A-H2B Histone Dimer. Biochemistry. 2002;41:14951–59. doi: 10.1021/bi026282s. [DOI] [PubMed] [Google Scholar]
- 68.Placek BJ, Harrison LN, Villers BM, Gloss LM. The H2A.Z/H2B dimer is unstable compared to the dimer containing the major H2A isoform. Protein Sci. 2005;14:514–22. doi: 10.1110/ps.041026405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nelson DA, Alonso WR. Extraction of histones H2A, H3 and H4 from yeast nuclei. Measurement of the extent of yeast histone acetylation following one-dimensional gel electrophoresis. Biochim Biophys Acta. 1983;741:269–71. doi: 10.1016/0167-4781(83)90068-4. [DOI] [PubMed] [Google Scholar]
- 70.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–26. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 71.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 72.Banks DD, Gloss LM. Folding mechanism of the (H3-H4)2 histone tetramer of the core nucleosome. Protein Sci. 2004;13:1304–16. doi: 10.1110/ps.03535504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–80. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 74.Santoro MM, Bolen DW. A test of the linear extrapolation of unfolding free energy changes over an extended denaturant concentration range. Biochemistry. 1992;31:4901–7. doi: 10.1021/bi00135a022. [DOI] [PubMed] [Google Scholar]
- 75.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: Relation to changes in accessble surface areas of protein folding. Protein Science. 1995;4:2138–48. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zitzewitz JA, Bilsel O, Luo J, Jones BE, Matthews CR. Probing the folding mechanism of a leucine zipper peptide by stopped-flow circular dichroism spectroscopy. Biochemistry. 1995;34:12812–19. doi: 10.1021/bi00039a042. [DOI] [PubMed] [Google Scholar]
- 77.Bilsel O, Zitzewitz JA, Bowers KE, Matthews CR. Folding mechanism of the alpha-subunit of tryptophan synthase, an alpha/beta barrel protein: global analysis highlights the interconversion of multiple native, intermediate, and unfolded forms through parallel channels. Biochemistry. 1999;38:1018–29. doi: 10.1021/bi982365q. [DOI] [PubMed] [Google Scholar]
- 78.Beechem JM. Global analysis of biochemical and biophysical data. Methods in Enzymology. 1992;210:37–54. doi: 10.1016/0076-6879(92)10004-w. [DOI] [PubMed] [Google Scholar]
- 79.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–50. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.