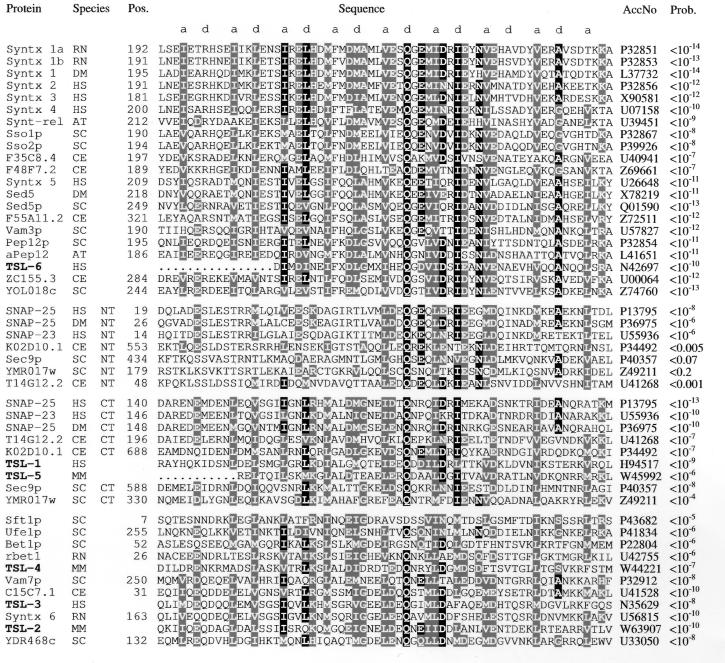
Figure 1.
Alignment of the coiled-coil t-SNARE homology domain. The header line shows the register of the predicted coiled-coil. The t-SNARE domains of representative members of the syntaxin family (first sequence block), the SNAP-25 family (second and third blocks), and members of other families or of uncertain family relation (fourth block) are shown with all newly identified mammalian homologous proteins (TSLs, boldface type). Residues conserved in more than 50% of all sequences are printed inversely; positions with conservative substitutions in more than 50% of all sequences are printed on a shaded background. Protein or gene names are indicated, and two-letter species abbreviations is as follows: RN, Rattus norvegicus; HS, Homo sapiens; DM, Drosophila melanogaster; CE, C. elegans; SC, Saccharomyces cerevisiae; AT, Arabidopsis thaliana; MM, Mus musculus. NT and CT refer to the N-terminal and C-terminal homology domain of SNAP-25-related proteins, respectively. Numbers to the left of the alignment indicate the position of the homology domain in the sequence (if known). The sequence accession numbers (AccNo) are shown. Codes starting with the letters P or Q are from SwissProt; others are from GenBank/EMBL. The statistical significance of the sequences matching to the final domain profile are expressed as error probabilities (Prob.). Each individual sequence was optimally aligned against the profile by using the pftools program package. Coiled-coil prediction with the program coils 2.1, using standard parameters (Window width, 21/28; MTIDK matrix) gave coiled-coil probabilities of P > 0.7 over a range of at least 35 residues for all domains listed with two exceptions. TSL-4 and YDR468c reached only maximal values of 0.45 and 0.3, respectively. If the problems in coiled-coil detection for short segments (26) are taken into account, it is likely that all domains adopt a coiled-coil conformation with similar geometry.