Abstract
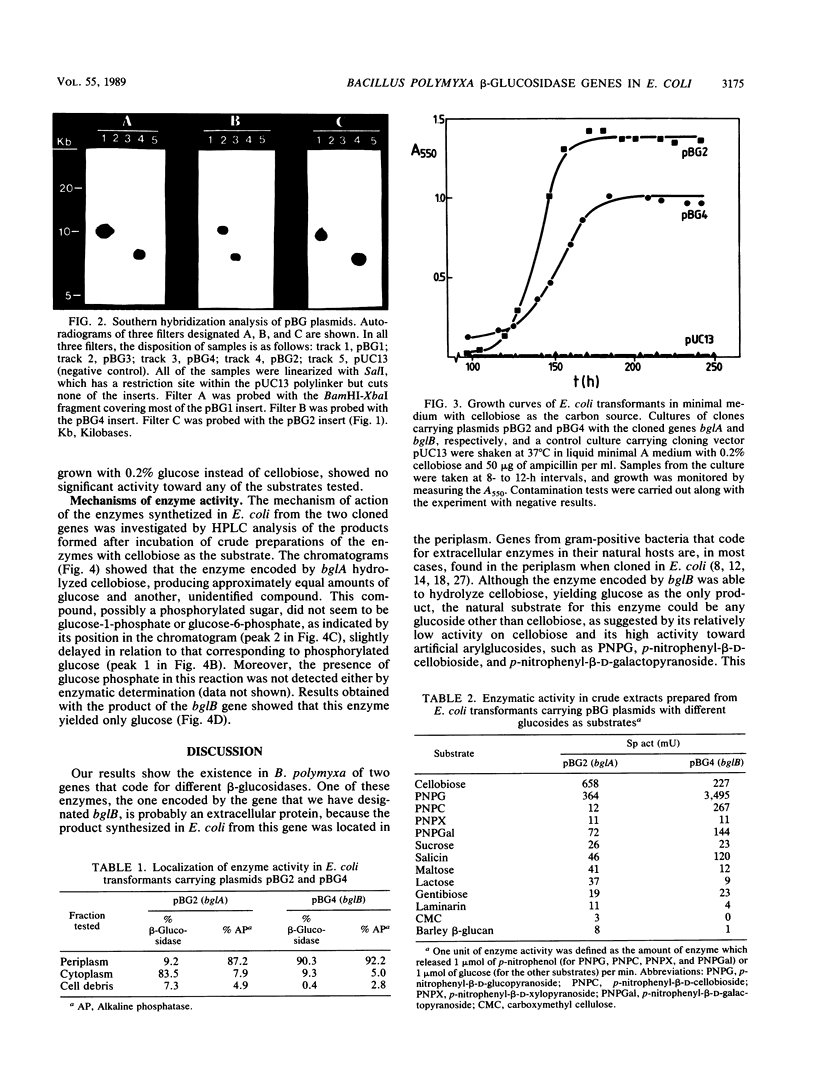
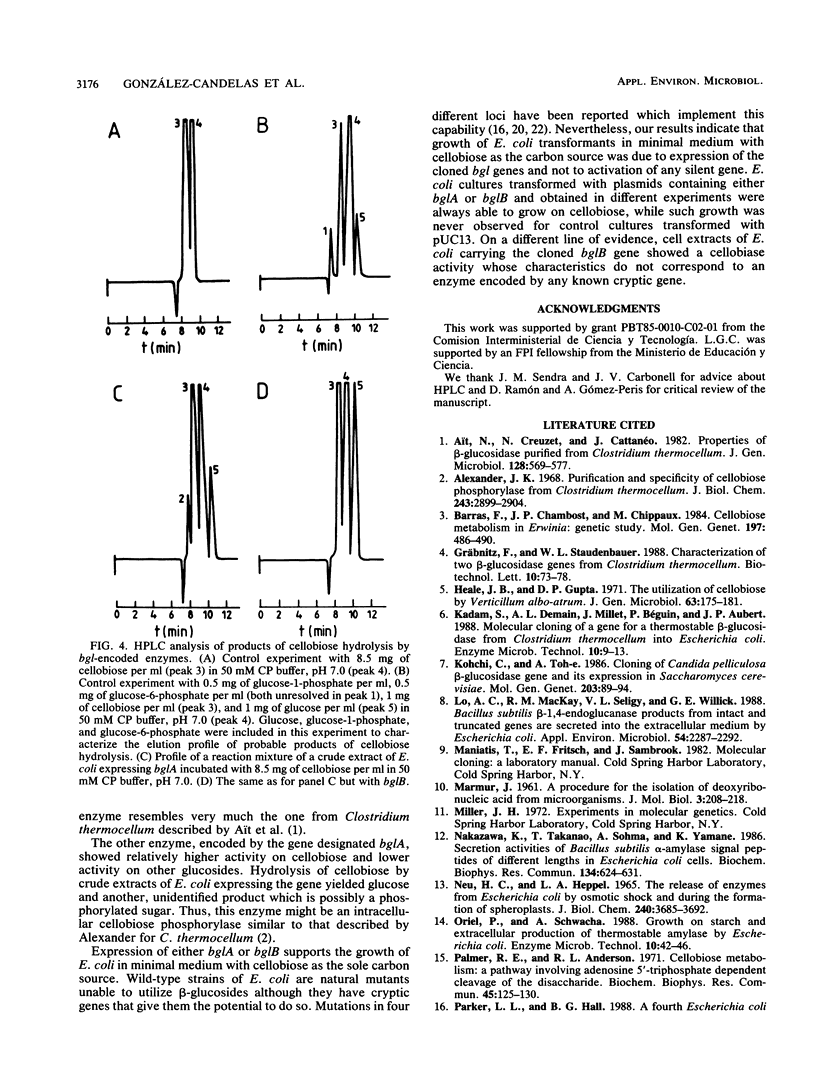
DNA fragments from Bacillus polymyxa which encode beta-glucosidase activity were cloned in Escherichia coli by selection of yellow transformants able to hydrolyze the artificial chromogenic substrate p-nitrophenyl-beta-D-glucopyranoside. Restriction endonuclease maps and Southern analysis of the cloned fragments showed the existence of two different genes. Expression of either one of these genes allowed growth of E. coli in minimal medium with cellobiose as the only carbon source. One of the two enzymes was found in the periplasm of E. coli, hydrolyzed arylglucosides more actively than cellobiose, and rendered glucose as the only product upon cellobiose hydrolysis. The other enzyme was located in the cytoplasm, was more active toward cellobiose, and hydrolyzed this disaccharide, yielding glucose and another, unidentified compound, probably a phosphorylated sugar.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander J. K. Purification and specificity of cellobiose phosphorylase from Clostridium thermocellum. J Biol Chem. 1968 Jun 10;243(11):2899–2904. [PubMed] [Google Scholar]
- Barras F., Chambost J. P., Chippaux M. Cellobiose metabolism in Erwinia: genetic study. Mol Gen Genet. 1984;197(3):486–490. doi: 10.1007/BF00329947. [DOI] [PubMed] [Google Scholar]
- Heale J. B., Gupta D. P. The utilization of cellobiose by Verticillium albo-atrum. J Gen Microbiol. 1970 Oct;63(2):175–181. doi: 10.1099/00221287-63-2-175. [DOI] [PubMed] [Google Scholar]
- Kohchi C., Toh-e A. Cloning of Candida pelliculosa beta-glucosidase gene and its expression in Saccharomyces cerevisiae. Mol Gen Genet. 1986 Apr;203(1):89–94. doi: 10.1007/BF00330388. [DOI] [PubMed] [Google Scholar]
- Lo A. C., MacKay R. M., Seligy V. L., Willick G. E. Bacillus subtilis beta-1,4-endoglucanase products from intact and truncated genes are secreted into the extracellular medium by Escherichia coli. Appl Environ Microbiol. 1988 Sep;54(9):2287–2292. doi: 10.1128/aem.54.9.2287-2292.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa K., Takano T., Sohma A., Yamane K. Secretion activities of Bacillus subtilis alpha-amylase signal peptides of different lengths in Escherichia coli cells. Biochem Biophys Res Commun. 1986 Jan 29;134(2):624–631. doi: 10.1016/s0006-291x(86)80465-x. [DOI] [PubMed] [Google Scholar]
- Neu H. C., Heppel L. A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965 Sep;240(9):3685–3692. [PubMed] [Google Scholar]
- Palmer R. E., Anderson R. L. Cellobiose metabolism: a pathway involving adenosine 5'-triphosphate-dependent cleavage of the disaccharide. Biochem Biophys Res Commun. 1971 Oct 1;45(1):125–130. doi: 10.1016/0006-291x(71)90059-3. [DOI] [PubMed] [Google Scholar]
- Parker L. L., Hall B. G. A fourth Escherichia coli gene system with the potential to evolve beta-glucoside utilization. Genetics. 1988 Jul;119(3):485–490. doi: 10.1093/genetics/119.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynal A., Guerineau M. Cloning and expression of the structural gene for beta-glucosidase of Kluyveromyces fragilis in Escherichia coli and Saccharomyces cerevisiae. Mol Gen Genet. 1984;195(1-2):108–115. doi: 10.1007/BF00332732. [DOI] [PubMed] [Google Scholar]
- Schaefler S. Inducible system for the utilization of beta-glucosides in Escherichia coli. I. Active transport and utilization of beta-glucosides. J Bacteriol. 1967 Jan;93(1):254–263. doi: 10.1128/jb.93.1.254-263.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz K., Toloczyki C., Rak B. Beta-glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J Bacteriol. 1987 Jun;169(6):2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewale J. G. Beta-Glucosidase: its role in cellulase synthesis and hydrolysis of cellulose. Int J Biochem. 1982;14(6):435–443. doi: 10.1016/0020-711x(82)90109-4. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yang R. C., Mackenzie C. R., Bilous D., Seligy V. L., Narang S. A. Molecular Cloning and Expression of a Xylanase Gene from Bacillus polymyxa in Escherichia coli. Appl Environ Microbiol. 1988 Apr;54(4):1023–1029. doi: 10.1128/aem.54.4.1023-1029.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]



