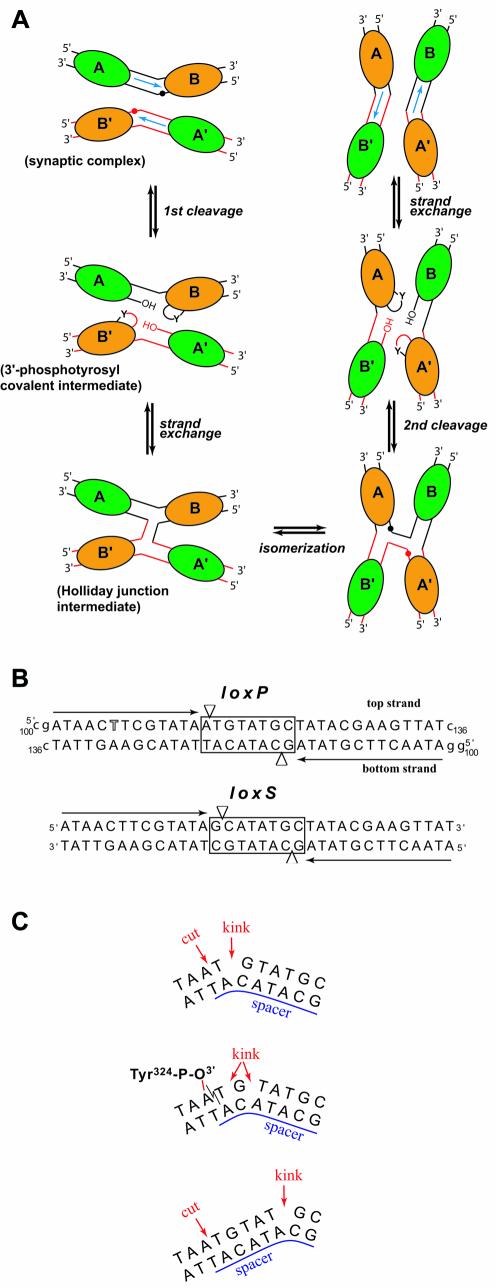
Figure 1.
(A) Schematic representation of Cre–loxP-mediated recombin ation. Cre molecules are represented by ellipses (green for non-cleaving subunits and orange for active subunits) and labeled A, B, A′ and B′. Red and black spheres indicate cleavage sites. This figure is adapted from Gopaul et al. (8). (B) Sequences of the loxP (used in this study) and the symmetrized loxS (used by Van Duyne’s group) DNA. The 13 bp inverted repeat sequences are indicated with black arrows. The thymidine position substituted by a 5-iodo-deoxyuridine is highlighted. The spacer region is boxed and arrowheads depict cleavage sites. Residues noted in lower case were added at the 3′ and 5′ ends for crystallization of the complex. (C) In the Cre–loxP synaptic complex, the 8 bp spacer region is kinked immediately next to the top strand cleavage site (top). After the first cleavage, a 3′-phosphotyrosine covalent intermediate is formed (middle). Some rearrangements in the spacer region redistribute the kink over several residues in the cleaved DNA but not all base pairs are still formed. Unlike in the present structures, in the Cre–loxS synaptic complex, the kink is 5 bp away from the cleavage site (bottom).