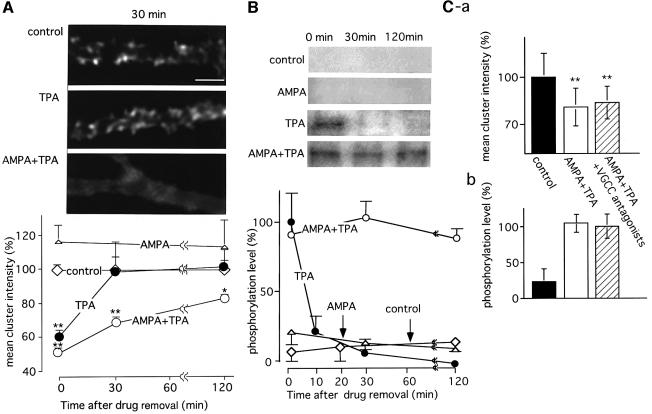
Fig. 4. Long-lasting disruption of GluR2 clusters in association with persistent phosphorylation of the Ser880 residue in GluR2 following conjunctive stimulation with TPA and AMPA. (A) Time course of changes in the density of GluR2 clusters. Cerebellar neuronal cultures were stimulated with TPA (200 nM, 20 min), AMPA (100 µM, 1 min) or TPA+AMPA, and incubated for 0, 30 and 120 min after the removal of the drugs. Then, the cells were fixed and immunostained for GluR2. Upper panels indicate the disruption of GluR2 immunoreactivity on Purkinje cell dendrites at 30 min after removal of the drugs (scale bar 5 µm). For quantitative analysis of the effect of TPA on GluR2 cluster density, the mean of the maximum cluster intensities was calculated from five different cells. The results are plotted on the graph, with the mean intensity in the control culture being 100%. (B) Time course of changes in Ser880 phosphorylation levels following various stimulations. Cerebellar neuronal cultures were treated as described above. After a set period of incubation, the cells were solubilized, and the membrane fractions were subjected to SDS–PAGE, followed by immunoblotting with anti-P-Ser880 antibodies (insets). The means of the band intensities from three independent experiments are plotted on the graph. The band intensities obtained from TPA-treated cultures without further incubation were defined as 100%. Error bars indicate SD. (C) No significant effect of VGCC blockers on TPA+AMPA induced disruption of the GluR2 clusters and phosphorylation of Ser880 in GluR2. TPA+AMPA stimulation was applied in the presence or absence of VGCC blockers, verapamil (10 µM) and ω-agatoxin TK (50 nM), and incubation was carried out for 30 min after the removal of drugs. Then, mean cluster intensity (a) and the phosphorylation level (b) of GluR2 were quantified. Asterisks indicate the significant difference as compared with control samples (*p <0.05, **p <0.01).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.