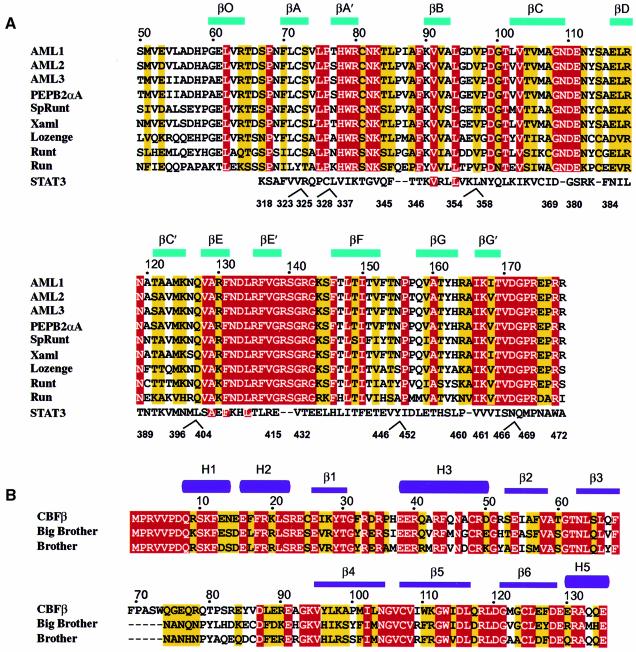
Fig. 3. Amino acid sequence alignment of the Runt domain and CBFβ. (A) The amino acid sequence accession numbers for the Swiss-Prot and DDBJ/EMBL/GenBank databases are given in parentheses. The sequence information is derived from human AML1 (Q01196), residues 50–178; human AML2 (Q13761), residues 54–182; human AML3 (Q08775), residues 102–220; murine PEBP2αA (Q08775, D14636), residues 50–178; sea-urchin SpRunt-1 (Q26628), residues 57–185; frog Xaml1 (O73725), residues 50–178; fruit-fly Lozenge (Q24183), residues 278–406 and Runt (Q24709), residues 106–234; nematode Run (O01834), residues 10–138. The last line of the alignment shows the structural similarity of the Runt domain to murine STAT3β (P42227), residues 318–472, with manually introduced sequence gaps indicated. Identical residues are highlighted in red, conservatively substituted residues are highlighted in yellow. Sequences were aligned using CLUSTAL_W (Thompson et al., 1994) and manually adjusted. (B) Sequence alignment of human CBFβ (Q13951), residues 1–135, and the fruit-fly homologues Brother (Q24039) and Big Brother (Q24040). Residues 70–74 in CBFβ have no equivalent in Drosophila, and are indicated by dashed lines.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.